5E5F
 
 | |
6O9G
 
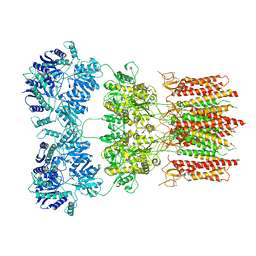 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
2GGH
 
 | |
3FTY
 
 | |
5FSO
 
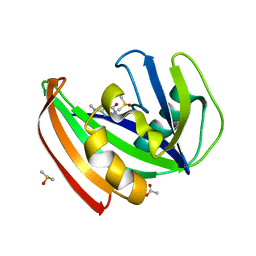 | | MTH1 substrate recognition: Complex with a methylaminopyrimidinedione. | | Descriptor: | 6-(METHYLAMINO)-1H-PYRIMIDINE-2,4-DIONE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | MTH1 Substrate Recognition--An Example of Specific Promiscuity.
PLoS ONE, 11, 2016
|
|
3MBP
 
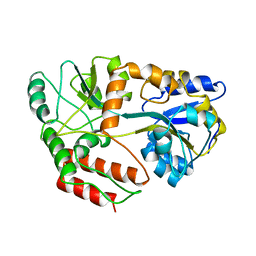 | | MALTODEXTRIN-BINDING PROTEIN WITH BOUND MALTOTRIOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
7D5C
 
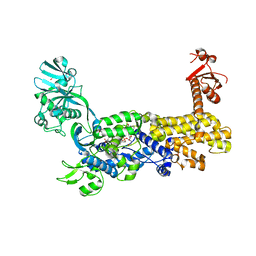 | | IleRS in complex with a tRNA site inhibitor | | Descriptor: | (2E,4S,5S,6E,8E)-10-[(2S,3R,6S,8R,9S)-3-butyl-9-methyl-2-[(1E,3E)-3-methyl-5-oxidanyl-5-oxidanylidene-penta-1,3-dienyl]-3-(4-oxidanyl-4-oxidanylidene-butanoyl)oxy-1,7-dioxaspiro[5.5]undecan-8-yl]-4,8-dimethyl-5-oxidanyl-deca-2,6,8-trienoic acid, 1,2-ETHANEDIOL, Isoleucine--tRNA ligase, ... | | Authors: | Chen, B, Luo, S, Zhou, H. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase.
Nat Commun, 12, 2021
|
|
3FUI
 
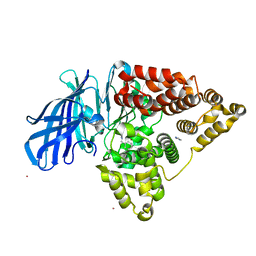 | |
1DPM
 
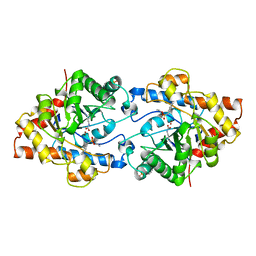 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIETHYL 4-METHYLBENZYLPHOSPHONATE | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, FORMIC ACID, PHOSPHOTRIESTERASE, ... | | Authors: | Vanhooke, J.L, Benning, M.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 1996-02-13 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
Biochemistry, 35, 1996
|
|
6DGJ
 
 | |
2GRH
 
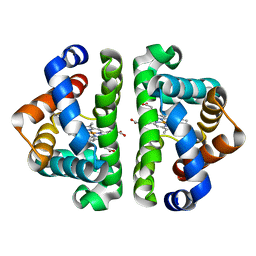 | | M37V mutant of Scapharca dimeric hemoglobin, with CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Pahl, R, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric action in real time: Time-resolved crystallographic studies of a cooperative dimeric hemoglobin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3FU5
 
 | |
5CID
 
 | |
2GGJ
 
 | |
3MVT
 
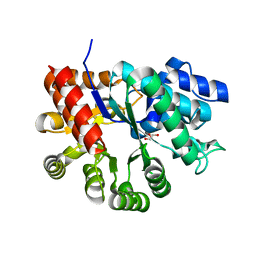 | | Crystal structure of apo mADA at 2.2A resolution | | Descriptor: | Adenosine deaminase, CHLORIDE ION, GLYCEROL | | Authors: | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
5SQH
 
 | |
5SQG
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894430 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5016127255 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SRW
 
 | |
5SSB
 
 | |
5SSP
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5459166285 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(1H-indole-5-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQE
 
 | |
5SRR
 
 | |
5SSC
 
 | |
5SRY
 
 | |
