5CI1
 
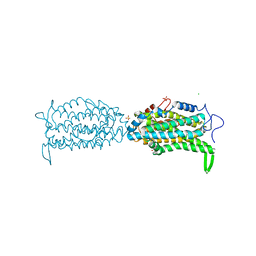 | | Ribonucleotide reductase Y122 2,3-F2Y variant | | Descriptor: | CHLORIDE ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
7PAC
 
 | | The crystal structure of PDE6D in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
6GBR
 
 | | Crystal Structure of the oligomerization domain of VP35 from Reston virus, mercury derivative | | Descriptor: | MERCURIBENZOIC ACID, Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
3FFW
 
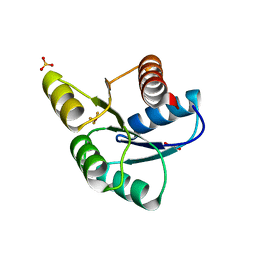 | | Crystal Structure of CheY triple mutant F14Q, N59K, E89Y complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
4KPL
 
 | | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg,d-mannonate and 2-keto-3-deoxy-d-gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-05-13 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg,d-mannonate and 2-keto-3-deoxy-d-gluconate
To be Published
|
|
3HY1
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with SerSA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXYETHYL DISULFIDE, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
1KO6
 
 | | Crystal Structure of C-terminal Autoproteolytic Domain of Nucleoporin Nup98 | | Descriptor: | Nuclear Pore Complex Protein Nup98 | | Authors: | Hodel, A.E, Hodel, M.R, Griffis, E.R, Hennig, K.A, Ratner, G.A, Songli, X, Powers, M.A. | | Deposit date: | 2001-12-20 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98.
Mol.Cell, 10, 2002
|
|
4TRZ
 
 | | Structure of BACE1 complex with 2-thiophenyl HEA-type inhibitor | | Descriptor: | 2-thiophenyl HEA-type inhibitor, Beta-secretase 1 | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
6E04
 
 | | sperm whale myoglobin 1-nitrosopropane | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
2B6E
 
 | | X-Ray Crystal Structure of Protein HI1161 from Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR63. | | Descriptor: | ACETIC ACID, Hypothetical UPF0152 protein HI1161 | | Authors: | Kuzin, A.P, Benach, J, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Cunningham, K.E, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the different crystal form of the hypothetical UPF0152 protein HI1161: NESG target IR63
TO BE PUBLISHED
|
|
4MW9
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1478) | | Descriptor: | 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MWB
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[(2,5-dichlorothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1509) | | Descriptor: | 1-(3-{[(2,5-dichlorothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
2OZQ
 
 | | Crystal Structure of apo-MUP | | Descriptor: | CADMIUM ION, Novel member of the major urinary protein (Mup) gene family, SODIUM ION | | Authors: | Dennis, C.A, Homans, S.W, Phillips, S.E.V, Syme, N.R. | | Deposit date: | 2007-02-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origin of heat capacity changes in a "nonclassical" hydrophobic interaction.
Chembiochem, 8, 2007
|
|
2WMF
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2OT5
 
 | |
5BK7
 
 | | The structure of MppP E15A mutant soaked with the substrate L-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
2P37
 
 | | Crystal structure of a lectin from Canavalia maritima seeds (CML) in complex with man1-3man-OMe | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Bezerra, G.A, Oliveira, T.M, Souza, E.P, Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W. | | Deposit date: | 2007-03-08 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
2WMD
 
 | | Crystal structure of NmrA-like family domain containing protein 1 in complex with NADP and 2-(4-chloro-phenylamino)-nicotinic acid | | Descriptor: | 2-(4-CHLORO-PHENYLAMINO)-NICOTINIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NMRA-LIKE FAMILY DOMAIN CONTAINING PROTEIN 1 | | Authors: | Bhatia, C, Pilka, E, Niesen, F, Yue, W.W, Ugochukwu, E, Savitsky, P, Hozjan, V, Roos, A.K, Filippakopoulos, P, von Delft, F, Heightman, T, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Nmra-Like Family Domain Containing Protein 1
To be Published
|
|
3HXZ
 
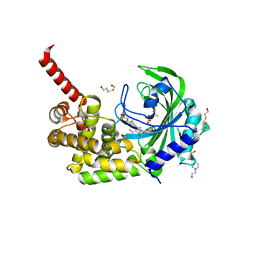 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
1JU7
 
 | |
4N49
 
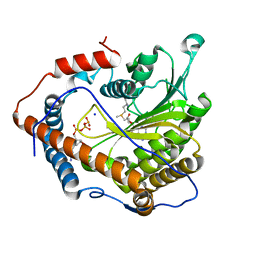 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
3OR1
 
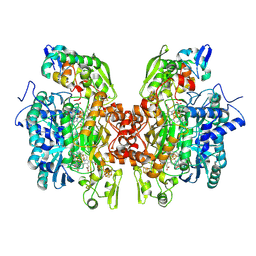 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
6E1N
 
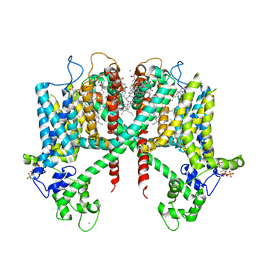 | | Structure of AtTPC1(DDE) in state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GIU
 
 | | Bianthranilate-like analogue bound in inner site of anthranilate phosphoribosyltransferase (AnPRT; trpD). | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-[(2-carboxy-5-methylphenyl)amino]-3-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-08-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
2P7V
 
 | | Crystal structure of the Escherichia coli regulator of sigma 70, Rsd, in complex with sigma 70 domain 4 | | Descriptor: | MAGNESIUM ION, RNA polymerase sigma factor rpoD, Regulator of sigma D | | Authors: | Patikoglou, G.A, Westblade, L.F, Campbell, E.A, Lamour, V, Lane, W.J, Darst, S.A. | | Deposit date: | 2007-03-20 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4.
J.Mol.Biol., 372, 2007
|
|
