8T11
 
 | |
1GQW
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Ryle, M.J, Clifton, I.J, Dunning-Hotopp, J.C, Lloyd, J.S, Burzlaff, N.I, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates
Biochemistry, 41, 2002
|
|
1H3B
 
 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-{6-[4-(6-BROMO-1,2-BENZISOTHIAZOL-3-YL)PHENOXY]HEXYL}-N-METHYL-2-PROPEN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H35
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4'-{[ALLYL(METHYL)AMINO]METHYL}-1,1'-BIPHENYL-4-YL)(4-BROMOPHENYL)METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
8TTH
 
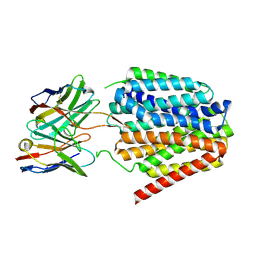 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
1H5V
 
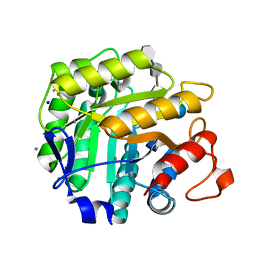 | | Thiopentasaccharide complex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, SODIUM ION, ... | | Authors: | Varrot, A, Sulzenbacher, G, Schulein, M, Driguez, H, Davies, G.J. | | Deposit date: | 2001-05-28 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Endoglucanase Cel5A in Complex with Methyl 4,4II,4III,4Iv-Tetrathio-Alpha-Cellopentoside Highlights the Alternative Binding Modes Targeted by Substrate Mimics
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8TPJ
 
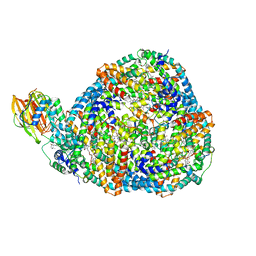 | | Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-04 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8USW
 
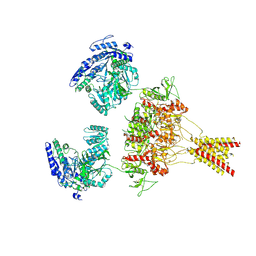 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
1HAB
 
 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
8TU1
 
 | |
1GNZ
 
 | | LECTIN I-B4 FROM GRIFFONIA SIMPLICIFOLIA (GS I-B4)METAL FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GSI-B4 ISOLECTIN, PHOSPHATE ION | | Authors: | Lescar, J, Loris, R, Mitchell, E, Gautier, C, Imberty, A. | | Deposit date: | 2001-10-11 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolectins I-A and I-B of Griffonia (Bandeiraea) Simplicifolia. Crystal Structure of Metal-Free Gs I-B(4) and Molecular Basis for Metal Binding and Monosaccharide Specificity.
J.Biol.Chem., 277, 2002
|
|
1HGU
 
 | | HUMAN GROWTH HORMONE | | Descriptor: | HUMAN GROWTH HORMONE | | Authors: | Chantalat, L, Jones, N, Korber, F, Navaza, J, Pavlovsky, A.G. | | Deposit date: | 1995-05-11 | | Release date: | 1995-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | THE CRYSTAL-STRUCTURE OF WILD-TYPE GROWTH-HORMONE AT 2.5 ANGSTROM RESOLUTION.
Protein Pept.Lett., 2, 1995
|
|
8U5B
 
 | |
1GW6
 
 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE D375N MUTANT | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Samuelsson, B, Haeggstrom, J.Z. | | Deposit date: | 2002-03-07 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Leukotriene A4 Hydrolase: Selective Abrogation of Leukotriene B4 Formation by Mutation of Aspartic Acid 375
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8U27
 
 | | Bcl-2-xL complexed with compound 35 | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-like protein 1 chimera, propan-2-yl {4-[(5S)-1-(4-bromobenzoyl)-5-phenyl-4,5-dihydro-1H-pyrazol-3-yl]phenyl}carbamate | | Authors: | Rizo, J, Pan, Y.-Z. | | Deposit date: | 2023-09-05 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights for selective disruption of Beclin 1 binding to Bcl-2.
Commun Biol, 6, 2023
|
|
1GWW
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
8TY1
 
 | |
8TZS
 
 | | Structure of human WLS | | Descriptor: | Protein wntless homolog | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZP
 
 | | Structure of human Wnt7a bound to WLS and RECK | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PALMITOLEIC ACID, Protein Wnt-7a, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8U5Y
 
 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
1HDK
 
 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
1HF6
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GZJ
 
 | |
1GZU
 
 | | Crystal Structure of Human Nicotinamide Mononucleotide Adenylyltransferase in Complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Werner, E, Ziegler, M, Lerner, F, Schweiger, M, Heinemann, U. | | Deposit date: | 2002-06-06 | | Release date: | 2002-07-05 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human nicotinamide mononucleotide adenylyltransferase in complex with NMN.
FEBS Lett., 516, 2002
|
|
1HBU
 
 | |
