2W17
 
 | | CDK2 in complex with the imidazole pyrimidine amide, compound (S)-8b | | Descriptor: | ACETATE ION, CELL DIVISION PROTEIN KINASE 2, N-(4-{[(3S)-3-(dimethylamino)pyrrolidin-1-yl]carbonyl}phenyl)-5-fluoro-4-[2-methyl-1-(1-methylethyl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Jones, C.D, Andrews, D.M, Barker, A.J, Blades, K, Daunt, P, East, S, Geh, C, Graham, M.A, Johnson, K.M, Loddick, S.A, McFarland, H.M, McGregor, A, Moss, L, Rudge, D.A, Simpson, P.B, Swain, M.L, Tam, K.Y, Tucker, J.A, Walker, M, Brassington, C, Haye, H, McCall, E. | | Deposit date: | 2008-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Discovery of Azd5597, a Potent Imidazole Pyrimidine Amide Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6QB7
 
 | | Structure of the H1 domain of human KCTD16 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, PHOSPHATE ION | | Authors: | Pinkas, D.M, Bufton, J.C, Williams, E.P, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the H1 domain of human KCTD16
To be published
|
|
6QS8
 
 | |
6QS4
 
 | |
6QS7
 
 | |
6QS6
 
 | |
6RN3
 
 | |
6S4L
 
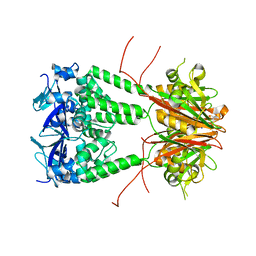 | | Structure of human KCTD1 | | Descriptor: | BTB/POZ domain-containing protein KCTD1, IODIDE ION, SODIUM ION | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Pike, A.C.W, Newman, J.A, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of human KCTD1
To be published
|
|
4B8X
 
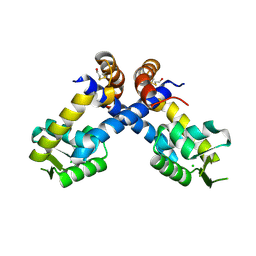 | | Near atomic resolution crystal structure of Sco5413, a MarR family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, POSSIBLE MARR-TRANSCRIPTIONAL REGULATOR | | Authors: | Holley, T.A, Stevenson, C.E.M, Bibb, M.J, Lawson, D.M. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Crystal Structure of Sco5413, a Widespread Actinomycete Marr Family Transcriptional Regulator of Unknown Function.
Proteins, 81, 2013
|
|
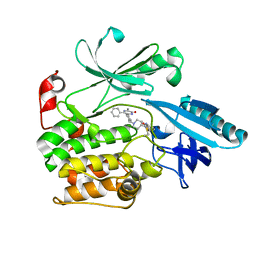
6S9W
 
 | |
6S9X
 
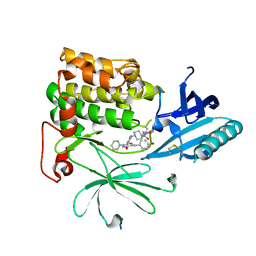 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 15c | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[3-[1-[[4-[5-[(4-hydroxyphenyl)methyl]-6-oxidanylidene-2-phenyl-1~{H}-pyrazin-3-yl]phenyl]methyl]piperidin-4-yl]-2-oxidanylidene-1~{H}-benzimidazol-5-yl]propanamide | | Authors: | Landel, I, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent-Allosteric Inhibitors to Achieve Akt Isoform-Selectivity.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6RN4
 
 | |
5U4I
 
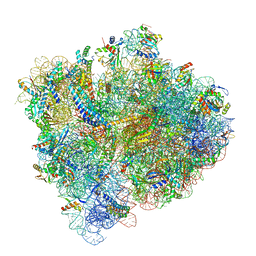 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
4CP4
 
 | |
6RN2
 
 | |
4CCX
 
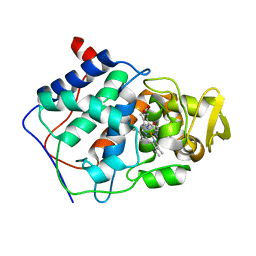 | | ALTERING SUBSTRATE SPECIFICITY AT THE HEME EDGE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-03-17 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Biochemistry, 35, 1996
|
|
2OQE
 
 | |
2OOV
 
 | |
4CHL
 
 | | Human Ethylmalonic Encephalopathy Protein 1 (hETHE1) | | Descriptor: | FE (III) ION, PERSULFIDE DIOXYGENASE ETHE1, MITOCHONDRIAL | | Authors: | Pettinati, I, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-12-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human persulfide dioxygenase: structural basis of ethylmalonic encephalopathy.
Hum. Mol. Genet., 24, 2015
|
|
7CZF
 
 | | Crystal structure of Kaposi Sarcoma associated herpesvirus (KSHV ) gHgL in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
5U4J
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
7BRI
 
 | |
7XBD
 
 | | Cryo-EM structure of human galanin receptor 2 | | Descriptor: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ishimoto, N, Kita, S, Park, S.Y. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
6DFP
 
 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6DWM
 
 | | Structure of Human Cytochrome P450 1A1 with Bergamottin | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 4-{[(2E)-3,7-dimethylocta-2,6-dien-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one, Cytochrome P450 1A1, ... | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands.
J. Biol. Chem., 293, 2018
|
|
