1WXH
 
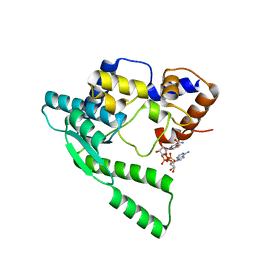 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1VCM
 
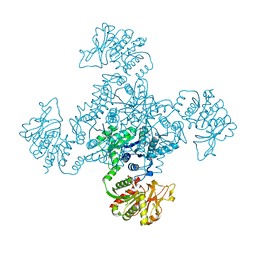 | |
1VK3
 
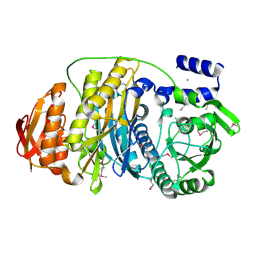 | |
1VCN
 
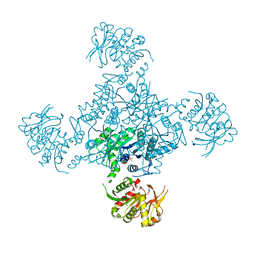 | |
1VQ3
 
 | |
1VCO
 
 | |
1WXE
 
 | | E.coli NAD Synthetase, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WV4
 
 | | X-ray Structure of Escherichia coli pyridoxine 5'-phosphate oxidase in tetragonal crystal form | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, Pyridoxamine 5'-phosphate oxidase | | Authors: | Safo, M.K, Musayev, F.N, Schirch, V. | | Deposit date: | 2004-12-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli pyridoxine 5'-phosphate oxidase in a tetragonal crystal form: insights into the mechanistic pathway of the enzyme.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WXF
 
 | | E.coli NAD Synthetase | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1UMY
 
 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
1XDJ
 
 | |
1TWJ
 
 | |
1TV5
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase with a bound inhibitor | | Descriptor: | (2Z)-2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]but-2-enamide, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, Dihydroorotate dehydrogenase homolog, ... | | Authors: | Hurt, D.E, Widom, J, Clardy, J. | | Deposit date: | 2004-06-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Plasmodium falciparum dihydroorotate dehydrogenase with a bound inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XMP
 
 | | Crystal Structure of PurE (BA0288) from Bacillus anthracis at 1.8 Resolution | | Descriptor: | phosphoribosylaminoimidazole carboxylase | | Authors: | Boyle, M.P, Kalliomaa, A.K, Levdikov, V, Blagova, E, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PurE (BA0288) from Bacillus anthracis at 1.8 A resolution
Proteins, 61, 2005
|
|
1XBV
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBX
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, 5-O-phosphono-L-ribulose, MAGNESIUM ION, ... | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XPG
 
 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Methyltetrahydrofolate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, MESO-ERYTHRITOL, ... | | Authors: | Pejchal, R, Ludwig, M.L. | | Deposit date: | 2004-10-08 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication
Plos Biol., 3, 2005
|
|
1U1J
 
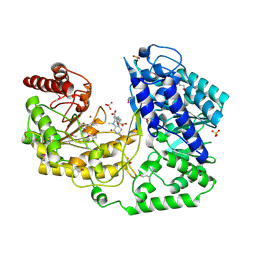 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
1U11
 
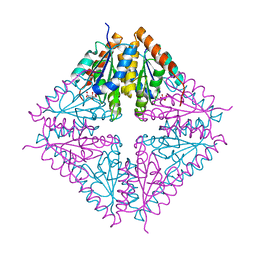 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U1U
 
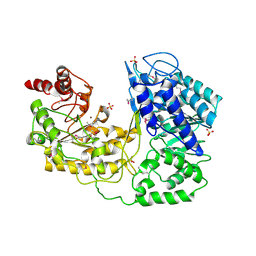 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, SULFATE ION, ZINC ION | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
1XR2
 
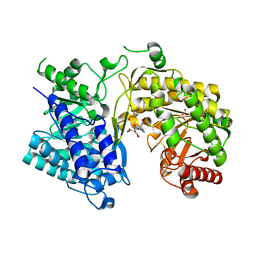 | | Crystal Structure of oxidized T. maritima Cobalamin-Independent Methionine Synthase complexed with Methyltetrahydrofolate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, MESO-ERYTHRITOL, ... | | Authors: | Pejchal, R, Ludwig, M.L. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication
Plos Biol., 3, 2005
|
|
6YF1
 
 | | FKBP12 in complex with the BMP potentiator compound 8 at 1.12A resolution | | Descriptor: | (1aR,3R,5S,6R,7S,9R,10R,17aS,20S,21R,22S,25R,25aR)-25-Ethyl-10,22-dihydroxy-20-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-5,7-dimethoxy-1a,3,9,21-tetramethyloctadecahydro-2H-6,10-epoxyoxireno[p]pyrido[2,1-c][1,4]oxazacyclotricosine-11,12,18,24(1aH,14H)-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF2
 
 | | FKBP12 in complex with the BMP potentiator compound 6 at 1.03A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-23,25-dimethoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14-bis(oxidanyl)-17-(2-oxidanylidenepropyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
