2CMC
 
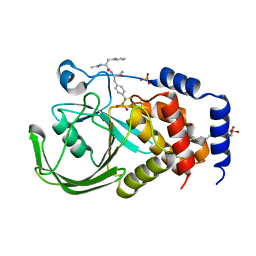 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-ACETYL-L-PHENYLALANYL-4-[DIFLUORO(PHOSPHONO)METHYL]-L-PHENYLALANINAMIDE, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, ... | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
1URJ
 
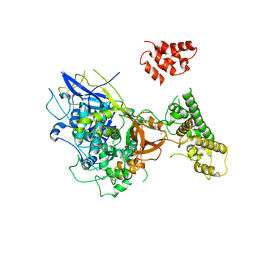 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | Descriptor: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Panjikar, S, Mapelli, M, Tucker, P.A. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
2CNF
 
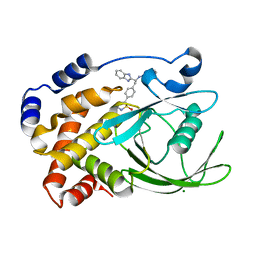 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | (5S)-5-{4-[(2S)-2-(1H-BENZIMIDAZOL-2-YL)-2-(1,3-BENZOTHIAZOL-2-YLAMINO)ETHYL]PHENYL}ISOTHIAZOLIDIN-3-ONE 1,1-DIOXIDE, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
1UTU
 
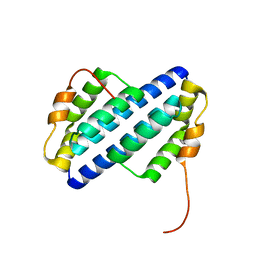 | |
1UW7
 
 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
1ZOP
 
 | | CD11A I-DOMAIN WITH BOUND MAGNESIUM ION | | Descriptor: | CHLORIDE ION, I-DOMAIN FRAGMENT OF LFA-1, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
2CSX
 
 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) | | Descriptor: | Methionyl-tRNA synthetase, RNA (75-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1ZID
 
 | |
1VBA
 
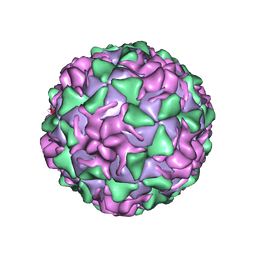 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
2CT8
 
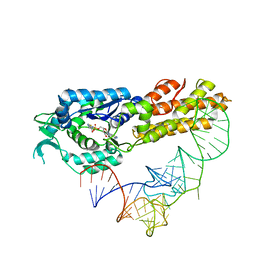 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) and methionyl-adenylate anologue | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, RNA (74-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YVS
 
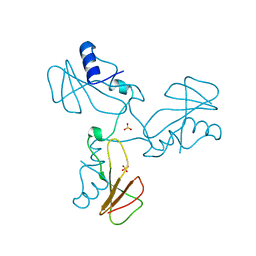 | | Trimeric domain swapped barnase | | Descriptor: | BARNASE, SULFATE ION | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1998-12-10 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric domain-swapped barnase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2CW7
 
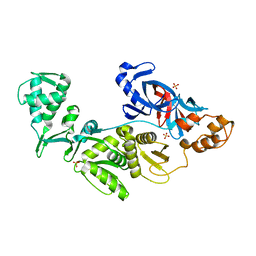 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
1YVE
 
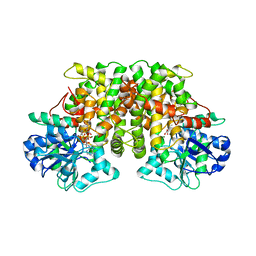 | | ACETOHYDROXY ACID ISOMEROREDUCTASE COMPLEXED WITH NADPH, MAGNESIUM AND INHIBITOR IPOHA (N-HYDROXY-N-ISOPROPYLOXAMATE) | | Descriptor: | ACETOHYDROXY ACID ISOMEROREDUCTASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Biou, V, Dumas, R, Cohen-Addad, C, Douce, R, Job, D, Pebay-Peyroula, E. | | Deposit date: | 1996-10-11 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of plant acetohydroxy acid isomeroreductase complexed with NADPH, two magnesium ions and a herbicidal transition state analog determined at 1.65 A resolution.
EMBO J., 16, 1997
|
|
1D5M
 
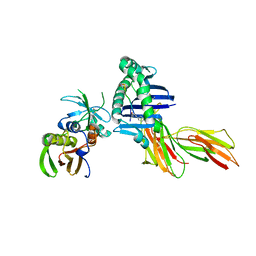 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDE AND SEB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Swain, A.L, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-07 | | Release date: | 2000-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1VAC
 
 | |
1ZON
 
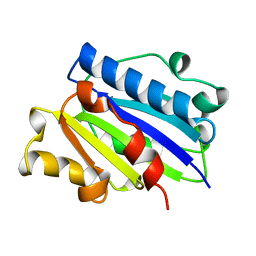 | | CD11A I-DOMAIN WITHOUT BOUND CATION | | Descriptor: | LEUKOCYTE ADHESION GLYCOPROTEIN | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-20 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
1VAD
 
 | |
1UPK
 
 | | Crystal structure of MO25 in complex with a C-terminal peptide of STRAD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MO25 PROTEIN, STE-20 RELATED ADAPTOR | | Authors: | Milburn, C.C, Boudeau, J, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Mo25 Alpha in Complex with the C-Terminus of the Pseudo Kinase Ste-20 Related Adaptor (Strad)
Nat.Struct.Mol.Biol., 11, 2004
|
|
1ZOO
 
 | | CD11A I-DOMAIN WITH BOUND MAGNESIUM ION | | Descriptor: | CHLORIDE ION, LEUKOCYTE ADHESION GLYCOPROTEIN, MAGNESIUM ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
1UR3
 
 | |
2CTS
 
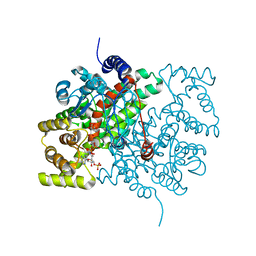 | |
1ZAK
 
 | |
1UKZ
 
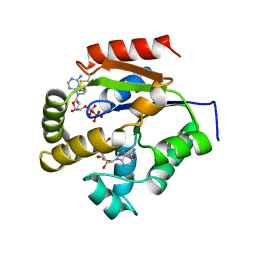 | |
1ZEN
 
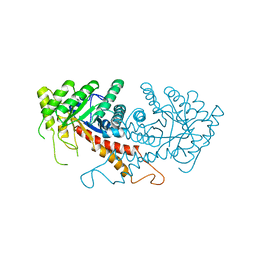 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Descriptor: | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE, ZINC ION | | Authors: | Cooper, S.J, Leonard, G.A, Hunter, W.N. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
Structure, 4, 1996
|
|
1UTM
 
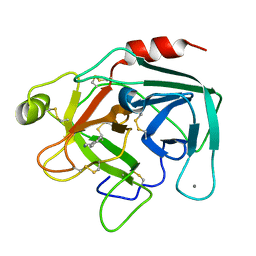 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, TRYPSIN I | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
