3W4I
 
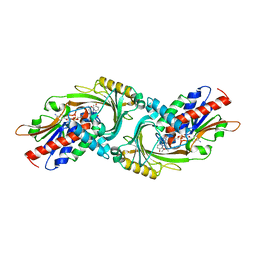 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
2IA8
 
 | | Kinetic and Crystallographic Studies of a Redesigned Manganese-Binding Site in Cytochrome c Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pfister, T, Mirarefi, A.Y, Gengenbach, A.J, Zhao, X, Conaster, C.D.N, Gao, Y.G, Robinson, H, Zukoski, C.F, Wang, A.H.J, Lu, Y. | | Deposit date: | 2006-09-07 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase
J.Biol.Inorg.Chem., 12, 2007
|
|
6TLJ
 
 | |
7LK3
 
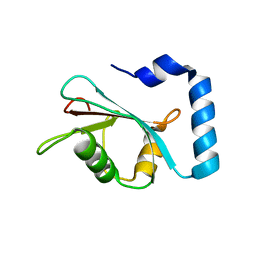 | | Crystal structure of untwinned human GABARAPL2 | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | Authors: | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1JTK
 
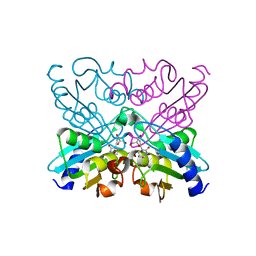 | | Crystal structure of cytidine deaminase from Bacillus subtilis in complex with the inhibitor tetrahydrodeoxyuridine | | Descriptor: | TETRAHYDRODEOXYURIDINE, ZINC ION, cytidine deaminase | | Authors: | Johansson, E, Mejlhede, N, Neuhard, J, Larsen, S. | | Deposit date: | 2001-08-21 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 A resolution.
Biochemistry, 41, 2002
|
|
6DG8
 
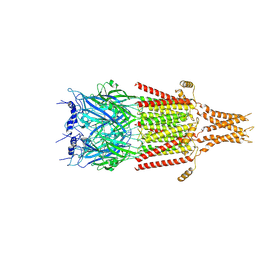 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
3FF9
 
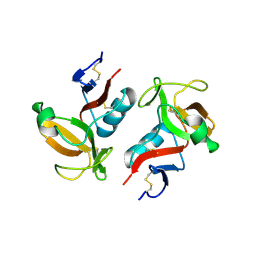 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
8FQX
 
 | | Carbonic Anhydrase II in complex with the alkyl ureas 3g | | Descriptor: | 4-hydroxy-3-({[(pyridin-4-yl)methyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FQZ
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3j | | Descriptor: | 4-hydroxy-3-{[(4-hydroxybutyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
5ZRN
 
 | |
8FR1
 
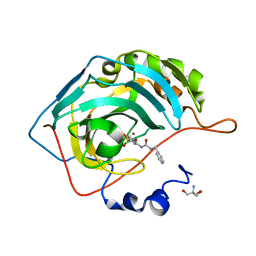 | | Carbonic Anhydrase IX in complex with the alkyl urea compound 3g | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-hydroxy-3-({[(pyridin-4-yl)methyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FQY
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3h | | Descriptor: | 4-hydroxy-3-({[2-(pyridin-2-yl)ethyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FR2
 
 | |
8FR4
 
 | |
6DG7
 
 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
4QH5
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) crystallized in phosphate buffer | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M, Sauguet, L. | | Deposit date: | 2014-05-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of potential modulation sites in the extracellular domain of the prokaryotic pentameric proton-gated ion channel GLIC
Acta Crystallogr.,Sect.D, 2015
|
|
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
1ZKC
 
 | | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | | Descriptor: | BETA-MERCAPTOETHANOL, Peptidyl-prolyl cis-trans isomerase like 2 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
1Y4T
 
 | | Ferric binding protein from Campylobacter jejuni | | Descriptor: | FE (III) ION, putative iron-uptake ABC transport system periplasmic iron-binding protein | | Authors: | Tom-Yew, S.A.L, Cui, D.T, Bekker, E.G, Murphy, M.E.P. | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anion-independent iron coordination by the Campylobacter jejuni ferric binding protein
J.Biol.Chem., 280, 2005
|
|
4I7E
 
 | | Crystal Structure of the Bacillus stearothermophilus Phosphofructokinase Mutant D12A in Complex with PEP | | Descriptor: | 6-phosphofructokinase, PHOSPHOENOLPYRUVATE | | Authors: | Mosser, R, Reddy, M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redefining the Role of the Quaternary Shift in Bacillus stearothermophilus Phosphofructokinase.
Biochemistry, 52, 2013
|
|
1MEZ
 
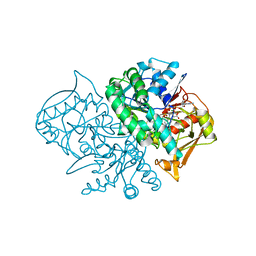 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1YBG
 
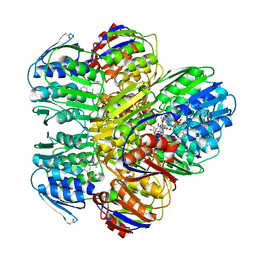 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
4XQM
 
 | |
1MF1
 
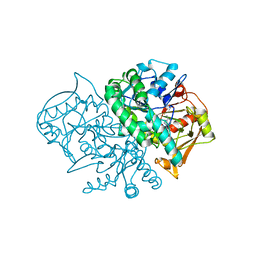 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
