4C4I
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(dimethylcarbamoyl)phenyl]amino}-2-(1,3-oxazol-5-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
1JDR
 
 | | Crystal Structure of a Proximal Domain Potassium Binding Variant of Cytochrome c Peroxidase | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Sundaramoorthy, M, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2001-06-14 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The effects of an engineered cation site on the structure, activity, and EPR properties of cytochrome c peroxidase.
Biochemistry, 38, 1999
|
|
1MK8
 
 | | Crystal Structure of a Mutant Cytochrome c Peroxidase showing a Novel Trp-Tyr Covalent Cross-link | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
1MKQ
 
 | | Crystal Structure of the Mutant Variant of Cytochrome c Peroxidase in the 'Open' Uncross-linked form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
3KQH
 
 | |
3KQL
 
 | |
3DTU
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
4QO5
 
 | | Hypothetical multiheme protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEME C, ... | | Authors: | Rajendran, C. | | Deposit date: | 2014-06-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | In meso crystal structure of a novel membrane-associated octaheme cytochrome c from the Crenarchaeon Ignicoccus hospitalis.
Febs J., 283, 2016
|
|
4RA5
 
 | | Human Protein Kinase C THETA IN COMPLEX WITH LIGAND COMPOUND 11a (6-[(1,3-Dimethyl-azetidin-3-yl)-methyl-amino]-4(R)-methyl-7-phenyl-2,10-dihydro-9-oxa-1,2,4a-triaza-phenanthren-3-one) | | Descriptor: | (1R)-9-[(1,3-dimethylazetidin-3-yl)(methyl)amino]-1-methyl-8-phenyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, 1,2-ETHANEDIOL, HUMAN PROTEIN KINASE C THETA, ... | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimized Protein Kinase C theta (PKC theta ) Inhibitors Reveal Only Modest Anti-inflammatory Efficacy in a Rodent Model of Arthritis.
J.Med.Chem., 58, 2015
|
|
5YRY
 
 | |
1ML2
 
 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase with Zn(II)-(20-oxo-Protoporphyrin IX) | | Descriptor: | 20-OXO-PROTOPORPHYRIN IX CONTAINING ZN(II), Cytochrome c Peroxidase | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
1LM2
 
 | | NMR structural characterization of the reduction of chromium(VI) to chromium(III) by cytochrome c7 | | Descriptor: | CHROMIUM ION, HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Bruschi, M, Michel, C, Turano, P. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The metal reductase activity of some multiheme cytochromes c: NMR structural characterization of the reduction of chromium(VI) to chromium(III) by cytochrome c(7).
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3BIZ
 
 | | Wee1 kinase complex with inhibitor PD331618 | | Descriptor: | 4-(2-chlorophenyl)-8-[3-(dimethylamino)propoxy]-9-hydroxy-6-methylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-dione, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-12-02 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3M97
 
 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4JIK
 
 | |
3FYI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides in the reduced state bound with cyanide | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
5O10
 
 | | Y48H mutant of human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Moreno-Chicano, T, Deacon, O.M, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Heightened Dynamics of the Oxidized Y48H Variant of Human Cytochrome c Increases Its Peroxidatic Activity.
Biochemistry, 56, 2017
|
|
4RA4
 
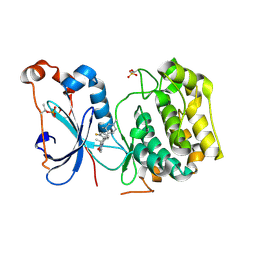 | | Crystal Structure of Human Protein Kinase C Alpha in Complex with Compound 28 ((R)-6-((3S,4S)-1,3-Dimethyl-piperidin-4-yl)-7-(2-fluoro-phenyl)-4-methyl-2,10-dihydro-9-oxa-1,2,4a-triaza-phenanthren-3-one) | | Descriptor: | (1R)-9-[(3S,4S)-1,3-dimethylpiperidin-4-yl]-8-(2-fluorophenyl)-1-methyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, Protein kinase C | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Optimized Protein Kinase C theta (PKC theta ) Inhibitors Reveal Only Modest Anti-inflammatory Efficacy in a Rodent Model of Arthritis.
J.Med.Chem., 58, 2015
|
|
1S05
 
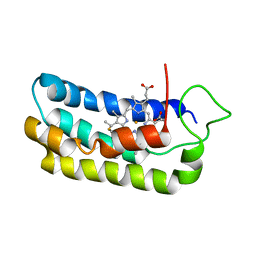 | | NMR-validated structural model for oxidized R.palustris cytochrome c556 | | Descriptor: | Cytochrome c-556, HEME C | | Authors: | Bertini, I, Faraone-Mennella, J, Gray, H.B, Luchinat, C, Parigi, G, Winkler, J.R. | | Deposit date: | 2003-12-30 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c(556).
J.Biol.Inorg.Chem., 9, 2004
|
|
2NRR
 
 | |
2NRX
 
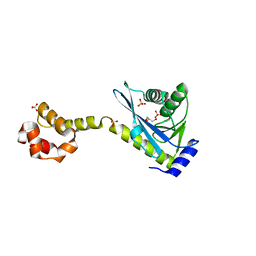 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | Descriptor: | GLYCEROL, SULFATE ION, UvrABC system protein C | | Authors: | Karakas, E, Truglio, J.J, Kisker, C. | | Deposit date: | 2006-11-02 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
6CI0
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with E101A (II) mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain.
Biochim. Biophys. Acta, 1859, 2018
|
|
1GX7
 
 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
7CRG
 
 | |
1N64
 
 | | Crystal structure analysis of the immunodominant antigenic site on Hepatitis C virus protein bound to mAb 19D9D6 | | Descriptor: | Fab 19D9D6 heavy chain, Fab 19D9D6 light chain, Genome polyprotein Capsid protein C | | Authors: | Menez, R, Bossus, M, Muller, B, Sibai, G, Dalbon, P, Ducancel, F, Jolivet-Reynaud, C, Stura, E. | | Deposit date: | 2002-11-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a hydrophobic immunodominant
antigenic site on hepatitis C virus core protein
complexed to monoclonal antibody 19D9D6.
J.Immunol., 170, 2003
|
|
