8SS2
 
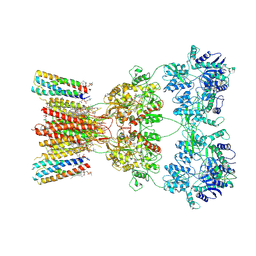 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SSA
 
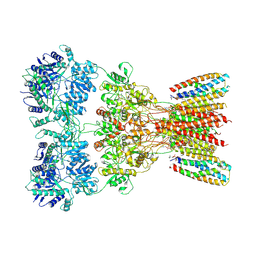 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to glutamate and channel blocker spermidine (desensitized state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS9
 
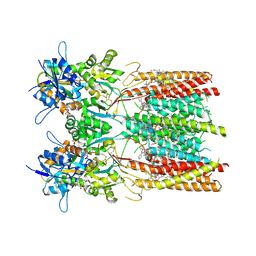 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS6
 
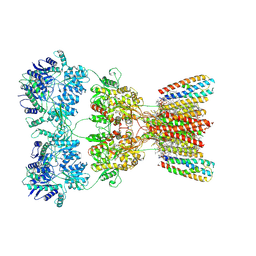 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS3
 
 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1YW0
 
 | | Crystal structure of the tryptophan 2,3-dioxygenase from Xanthomonas campestris. Northeast Structural Genomics Target XcR13. | | Descriptor: | MAGNESIUM ION, tryptophan 2,3-dioxygenase | | Authors: | Vorobiev, S.M, Abashidze, M, Forouhar, F, Kuzin, A, Xiao, R, Ciano, M, Aton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tryptophan 2,3-dioxygenase from Xanthomonas campestris. Northeast Structural Genomics Target XcR13.
To be Published
|
|
1YEY
 
 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
3EK7
 
 | | Calcium-saturated GCaMP2 dimer | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
3VRF
 
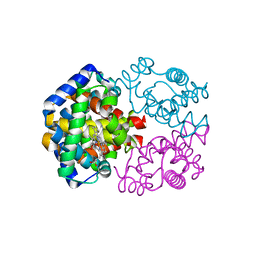 | | The crystal structure of hemoglobin from woolly mammoth in the carbonmonoxy forms | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3NDV
 
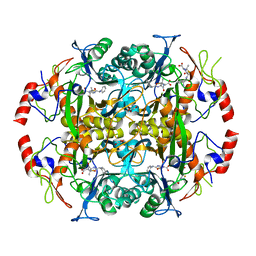 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with ampicillin | | Descriptor: | (2S,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
3EKH
 
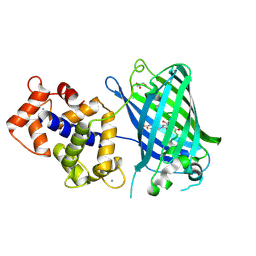 | | Calcium-saturated GCaMP2 T116V/K378W mutant monomer | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
3EK4
 
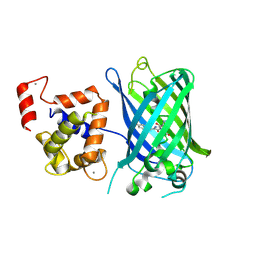 | | Calcium-saturated GCaMP2 Monomer | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
4U2P
 
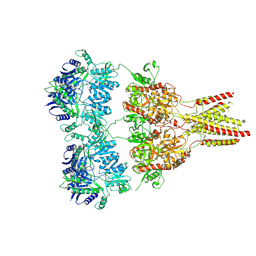 | |
1T86
 
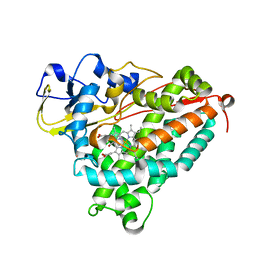 | | Crystal Structure of the Ferrous Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T85
 
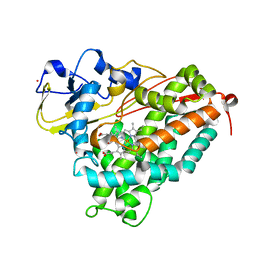 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, CARBON MONOXIDE, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
4U2Q
 
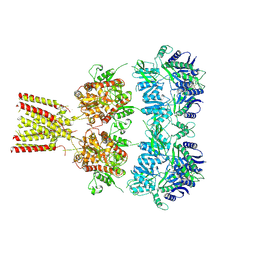 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5247 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
3UIB
 
 | | Map kinase LMAMPK10 from leishmania major in complex with SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, mitogen-activated protein kinase | | Authors: | Horjales, S, Schmidt-Arras, D, Leclercq, O, Spath, G, Buschiazzo, A. | | Deposit date: | 2011-11-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of the MAP Kinase LmaMPK10 from Leishmania Major Reveals Parasite-Specific Features and Regulatory Mechanisms.
Structure, 20, 2012
|
|
3BEP
 
 | | Structure of a sliding clamp on DNA | | Descriptor: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | Authors: | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
3VRE
 
 | | The crystal structure of hemoglobin from woolly mammoth in the deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VRG
 
 | | The crystal structure of hemoglobin from woolly mammoth in the met form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2QCS
 
 | | A complex structure between the Catalytic and Regulatory subunit of Protein Kinase A that represents the inhibited state | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kim, C, Cheng, C.Y, Saldanha, A.S, Taylor, S.S. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation.
Cell(Cambridge,Mass.), 130, 2007
|
|
6X0K
 
 | |
6AMS
 
 | |
2MBB
 
 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
6X0J
 
 | |
