2DZS
 
 | |
2DZW
 
 | |
2E09
 
 | |
1TAH
 
 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
1TCZ
 
 | |
2E08
 
 | |
1Y2G
 
 | | Crystal STructure of ZipA in complex with an inhibitor | | Descriptor: | Cell division protein zipA, N-METHYL-N-[3-(6-PHENYL[1,2,4]TRIAZOLO[4,3-B]PYRIDAZIN-3-YL)PHENYL]ACETAMIDE | | Authors: | Mosyak, L, Rush, T.S. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction
J.Med.Chem., 48, 2005
|
|
1Y0Q
 
 | | Crystal structure of an active group I ribozyme-product complex | | Descriptor: | 5'-R(*GP*CP*UP*U)-3', Group I ribozyme, MAGNESIUM ION, ... | | Authors: | Golden, B.L, Kim, H, Chase, E. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of a phage Twort group I ribozyme-product complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
1TKY
 
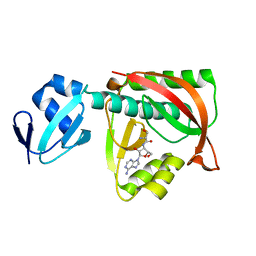 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1TL8
 
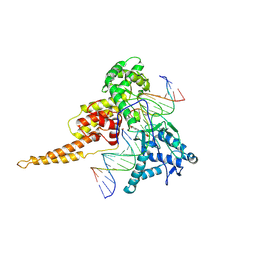 | | Human DNA topoisomerase I (70 kDa) in complex with the indenoisoquinoline AI-III-52 and covalent complex with a 22 base pair DNA duplex | | Descriptor: | 2,3-DIMETHOXY-12H-[1,3]DIOXOLO[5,6]INDENO[1,2-C]ISOQUINOLIN-6-IUM, 5'-D(*(TPC)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Ioanoviciu, A, Antony, S, Pommier, Y, Staker, B.L, Stewart, L, Cushman, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis and Mechanism of Action Studies of a Series of Norindenoisoquinoline Topoisomerase I Poisons Reveal an Inhibitor with a Flipped Orientation in the Ternary DNA-Enzyme-Inhibitor Complex As Determined by X-ray Crystallographic Analysis
J.Med.Chem., 48, 2005
|
|
1Y4M
 
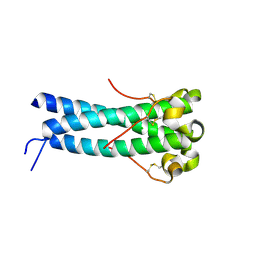 | | Crystal structure of human endogenous retrovirus HERV-FRD envelope protein (syncitin-2) | | Descriptor: | CHLORIDE ION, HERV-FRD_6p24.1 provirus ancestral Env polyprotein | | Authors: | Renard, M, Varela, P.F, Letzelter, C, Duquerroy, S, Rey, F.A, Heidmann, T. | | Deposit date: | 2004-12-01 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a pivotal domain of human syncytin-2, a 40 million years old endogenous retrovirus fusogenic envelope gene captured by primates.
J.Mol.Biol., 352, 2005
|
|
2E40
 
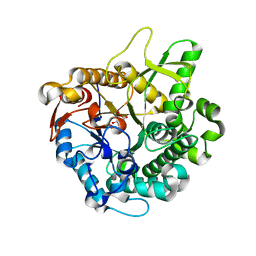 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
1Y3N
 
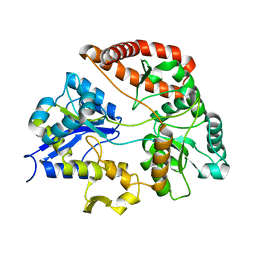 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1T39
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE COVALENTLY CROSSLINKED TO DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(E1X)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Y44
 
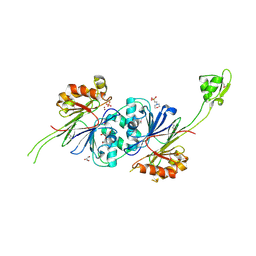 | | Crystal structure of RNase Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | de la Sierra-Gallay, I.L, Pellegrini, O, Condon, C. | | Deposit date: | 2004-11-30 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature, 433, 2005
|
|
2ECK
 
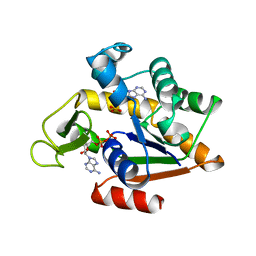 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
1T8F
 
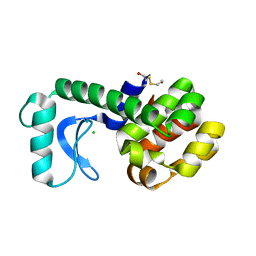 | | Crystal structure of phage T4 lysozyme mutant R14A/K16A/I17A/K19A/T21A/E22A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
1T8M
 
 | | CRYSTAL STRUCTURE OF THE P1 HIS BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-13 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
1AVO
 
 | | PROTEASOME ACTIVATOR REG(ALPHA) | | Descriptor: | 11S REGULATOR | | Authors: | Hill, C.P, Knowlton, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the proteasome activator REGalpha (PA28alpha).
Nature, 390, 1997
|
|
2EFQ
 
 | | Crystal Structure of Thr134 to Ala of ST1022-Glutamine Complex from Sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding.
Nucleic Acids Res., 36, 2008
|
|
2EG3
 
 | | Crystal Structure of Probable Thiosulfate Sulfurtransferase | | Descriptor: | Probable thiosulfate sulfurtransferase, SULFATE ION, ZINC ION | | Authors: | Sakai, H, Ebihara, A, Kitamura, Y, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Probable Thiosulfate Sulfurtransferase
To be Published
|
|
1ZG7
 
 | | Crystal Structure of 2-(5-{[amino(imino)methyl]amino}-2-chlorophenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 2-(5-{[AMINO(IMINO)METHYL]AMINO}-2-CHLOROPHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1T97
 
 | |
2E7Q
 
 | | Crystal structure of basic winged bean lectin in complex with b blood group trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Generation of blood group specificity: New insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin
Proteins, 68, 2007
|
|
1TAL
 
 | | ALPHA-LYTIC PROTEASE AT 120 K (SINGLE STRUCTURE MODEL) | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Rader, S.D, Agard, D.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational substates in enzyme mechanism: the 120 K structure of alpha-lytic protease at 1.5 A resolution.
Protein Sci., 6, 1997
|
|
