4WRO
 
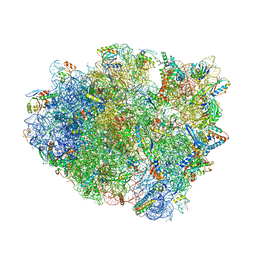 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
2X0V
 
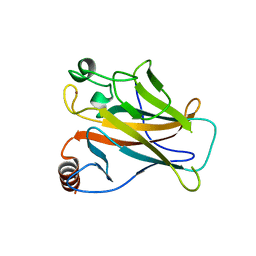 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2X0W
 
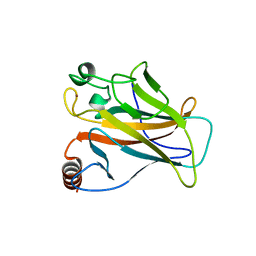 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 5,6-dimethoxy- 2-methylbenzothiazole | | Descriptor: | 5,6-DIMETHOXY-2-METHYL-1,3-BENZOTHIAZOLE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Kaar, J.L, Basse, N, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5LZX
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a UGA stop codon. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
2X0U
 
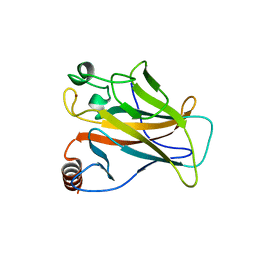 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
1JYE
 
 | | Structure of a Dimeric Lac Repressor with C-terminal Deletion and K84L Substitution | | Descriptor: | GLYCEROL, Lactose Operon Repressor | | Authors: | Bell, C.E, Barry, J, Matthews, K.S, Lewis, M. | | Deposit date: | 2001-09-12 | | Release date: | 2001-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator.
J.Mol.Biol., 313, 2001
|
|
1JYF
 
 | | Structure of the Dimeric Lac Repressor with an 11-residue C-terminal Deletion. | | Descriptor: | GLYCEROL, Lactose Operon Repressor | | Authors: | Bell, C.E, Barry, J, Matthews, K.S, Lewis, M. | | Deposit date: | 2001-09-12 | | Release date: | 2001-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator.
J.Mol.Biol., 313, 2001
|
|
2IT9
 
 | |
2IKK
 
 | | Structural Genomics, the crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Hypothetical transcriptional regulator yurK, SULFATE ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
6Z3R
 
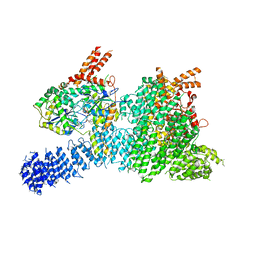 | | Structure of SMG1-8-9 kinase complex bound to UPF1-LSQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Gat, Y, Conti, E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity.
Elife, 9, 2020
|
|
6NT9
 
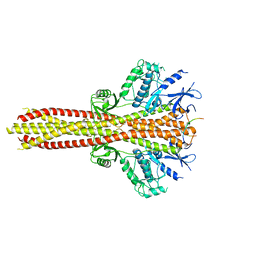 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
5WF0
 
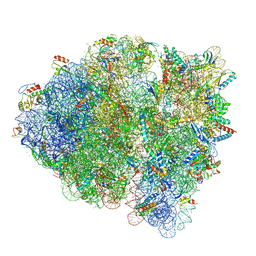 | |
5WFK
 
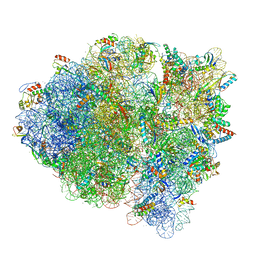 | |
6DD6
 
 | |
6D7N
 
 | |
5MHQ
 
 | | CCT068127 in complex with CDK2 | | Descriptor: | (2~{R},3~{S})-3-[[9-propan-2-yl-6-(pyridin-3-ylmethylamino)purin-2-yl]amino]pentan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Whittaker, S.R, Barlow, C, Martin, M.P, Mancusi, C, Wagner, S, Barrie, E, te Poele, R, Sharp, S, Brown, N, Wilson, S, Clarke, P, Walton, M.I, MacDonald, E, Blagg, J, Noble, M.E.M, Garrett, M.D, Workman, P. | | Deposit date: | 2016-11-25 | | Release date: | 2017-12-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor.
Mol Oncol, 12, 2018
|
|
5NEV
 
 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
3D0S
 
 | |
5WE4
 
 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WDT
 
 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
9F99
 
 | | Crystal structure of MUS81-EME1 bound by compound 10. | | Descriptor: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9M
 
 | | Crystal structure of MUS81-EME1 bound by compound 21. | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-(4-piperazin-1-ylphenyl)pyridine-3-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9K
 
 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9A
 
 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
