8E4W
 
 | |
5EQV
 
 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
2VXO
 
 | | Human GMP synthetase in complex with XMP | | Descriptor: | GMP SYNTHASE [GLUTAMINE-HYDROLYZING], SULFATE ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Welin, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-07-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity and Oligomerization of Human Gmp Synthetase
J.Mol.Biol., 425, 2013
|
|
7MR0
 
 | |
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
1L5R
 
 | | Human liver glycogen phosphorylase a complexed with riboflavin, N-Acetyl-beta-D-Glucopyranosylamine and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, N-acetyl-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
8EY9
 
 | | Structure of Arabidopsis fatty acid amide hydrolase mutant S305A in complex with 9-hydroxy-10,12-octadecadienoyl-ethanolamide | | Descriptor: | (9R,10E,12Z)-9-hydroxy-N-(2-hydroxyethyl)octadeca-10,12-dienamide, Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Gaguancela, O.A, Chapman, K.D. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural interactions explain the versatility of FAAH in the hydrolysis of plant and microbial acyl amide signals
To be published
|
|
5V4G
 
 | | Ruthenium(II)(cymene)(chlorido)2-lysozyme adduct with two binding sites | | Descriptor: | Lysozyme C, PARA-CYMENE RUTHENIUM CHLORIDE, SODIUM ION | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
8KGY
 
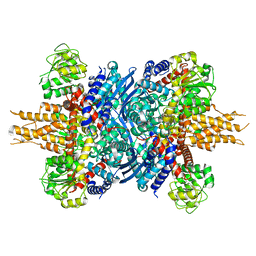 | | Human glutamate dehydrogenase I | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Su, M.-Y. | | Deposit date: | 2023-08-20 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8EWW
 
 | |
5F9X
 
 | | X-RAY STRUCTURE OF THE ADDUCT BETWEEN HEN EGG WHITE LYSOZYME AND CISPLATIN UPON 24 HOURS OF INCUBATION AT 55 DEGREES | | Descriptor: | Cisplatin, GLYCEROL, Lysozyme C | | Authors: | Russo Krauss, I, Ferraro, G, Pica, A, Merlino, A. | | Deposit date: | 2015-12-10 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Effect of temperature on the interaction of cisplatin with the model protein hen egg white lysozyme.
J.Biol.Inorg.Chem., 21, 2016
|
|
1G65
 
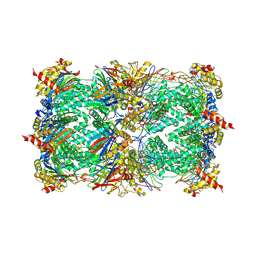 | | Crystal structure of epoxomicin:20s proteasome reveals a molecular basis for selectivity of alpha,beta-epoxyketone proteasome inhibitors | | Descriptor: | EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, Proteasome component C1, ... | | Authors: | Groll, M, Kim, K.B, Kairies, N, Huber, R, Crews, C. | | Deposit date: | 2000-11-03 | | Release date: | 2000-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Epoxomicin:20S Proteasome reveals a molecular basis for selectivity of alpha,beta-Epoxyketone Proteasome Inhibitors
J.Am.Chem.Soc., 122, 2000
|
|
6QWZ
 
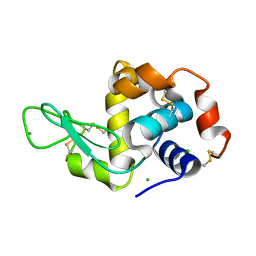 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
7MR2
 
 | |
5V5Z
 
 | | Structure of CYP51 from the pathogen Candida albicans | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Keniya, M.V, Sabherwal, M, Wilson, R.K, Sagatova, A.A, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2017-03-15 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Full-Length Lanosterol 14 alpha-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provide Tools for Antifungal Discovery.
Antimicrob.Agents Chemother., 62, 2018
|
|
7MR3
 
 | | Cryo-EM structure of RecBCD-DNA complex with docked RecBNuc and stabilized RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
2W0B
 
 | |
1RPN
 
 | | Crystal Structure of GDP-D-mannose 4,6-dehydratase in complexes with GDP and NADPH | | Descriptor: | GDP-mannose 4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Webb, N.A, Mulichak, A.M, Lam, J.S, Rocchetta, H.L, Garavito, R.M. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
Protein Sci., 13, 2004
|
|
6QY1
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 1500 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
8C8R
 
 | |
6HN2
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 5 | | Descriptor: | 14-3-3 protein sigma, 2-(3,4-dichlorophenyl)-~{N}-(2-sulfanylethyl)ethanamide, Estrogen Receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
5V7F
 
 | | T4 lysozyme Y18Ymi | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme | | Authors: | Carlsson, A.-C.C. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Increasing Enzyme Stability and Activity through Hydrogen Bond-Enhanced Halogen Bonds.
Biochemistry, 57, 2018
|
|
7MR4
 
 | | Cryo-EM structure of RecBCD-DNA complex with undocked RecBNuc and flexible RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
5V8X
 
 | |
5INB
 
 | | RepoMan-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
