6YQ6
 
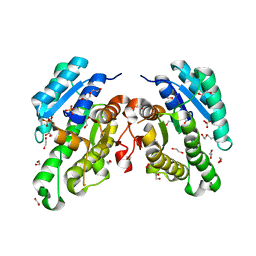 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
4QUU
 
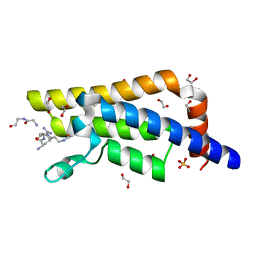 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) complexed with Histone H4-K(ac)5 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, Histone H4, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
J Mol Cell Biol, 8, 2016
|
|
1E3A
 
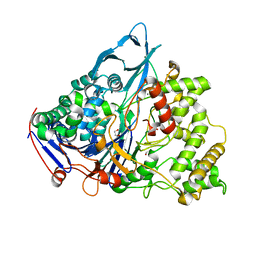 | | A slow processing precursor penicillin acylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hewitt, L, Kasche, V, Lummer, K, Lewis, R.J, Murshudov, G.N, Verma, C.S, Dodson, G.G, Wilson, K.S. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Slow Processing Precursor Penicillin Acylase from Escherichia Coli Reveals the Linker Peptide Blocking the Active-Site Cleft
J.Mol.Biol., 302, 2000
|
|
7OY2
 
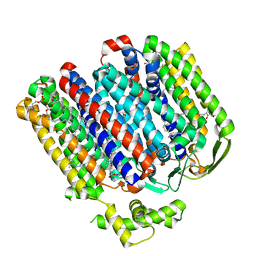 | | High resolution structure of cytochrome bd-II oxidase from E. coli | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(2~{E},6~{E},10~{Z},14~{E},18~{E},22~{E},26~{E})-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaenyl]naphthalene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Grund, T.N, Wu, D, Bald, D, Michel, H, Safarian, S. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Mechanistic and structural diversity between cytochrome bd isoforms of Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8FQZ
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3j | | Descriptor: | 4-hydroxy-3-{[(4-hydroxybutyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FQY
 
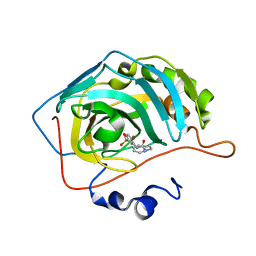 | | Carbonic Anhydrase II in complex with the alkyl urea 3h | | Descriptor: | 4-hydroxy-3-({[2-(pyridin-2-yl)ethyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8A2G
 
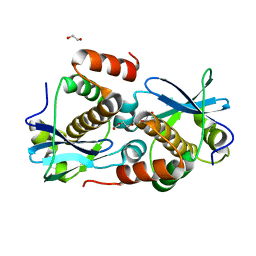 | | Crystal structure of Sebokelevirus 2A2 protein | | Descriptor: | 1,2-ETHANEDIOL, 2A2 protein, TETRAETHYLENE GLYCOL | | Authors: | Zhu, L, Von Castelmur, E, Whang, X, Ren, J, Fry, E, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
to be published
|
|
4RC5
 
 | |
4WID
 
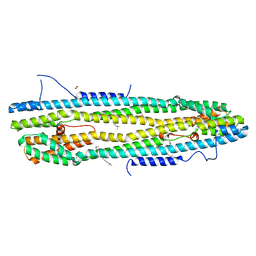 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
6Y6H
 
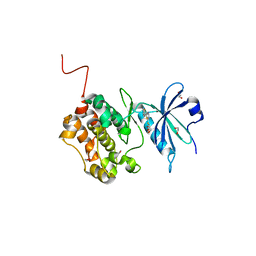 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
4WJ5
 
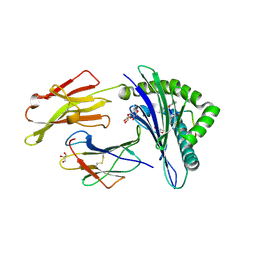 | | Structure of HLA-A2 in complex with an altered peptide ligands based on Mart-1 variant epitope | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Celie, P.H.N, Rodenko, B, Ovaa, H. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Altered Peptide Ligands Revisited: Vaccine Design through Chemically Modified HLA-A2-Restricted T Cell Epitopes.
J Immunol., 193, 2014
|
|
8G0C
 
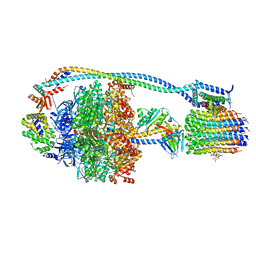 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
1DPO
 
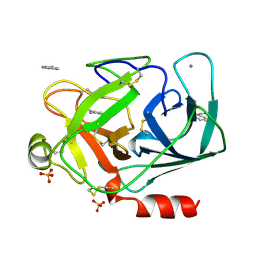 | | STRUCTURE OF RAT TRYPSIN | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stroud, R.M. | | Deposit date: | 1997-03-31 | | Release date: | 1997-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | 1.59 A structure of trypsin at 120 K: comparison of low temperature and room temperature structures.
Proteins, 10, 1991
|
|
6W0N
 
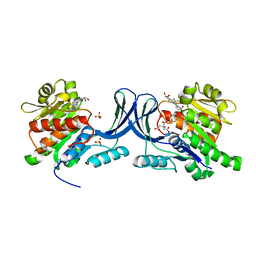 | | Structure of KHK in complex with compound 2 | | Descriptor: | 6-[(3~{S},4~{R})-3,4-bis(oxidanyl)pyrrolidin-1-yl]-2-[(3~{S})-3-methyl-3-oxidanyl-pyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-03-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of PF-06835919: A Potent Inhibitor of Ketohexokinase (KHK) for the Treatment of Metabolic Disorders Driven by the Overconsumption of Fructose.
J.Med.Chem., 63, 2020
|
|
4WIC
 
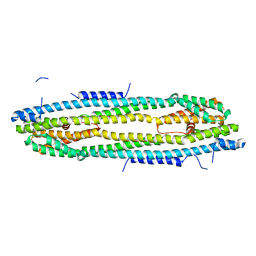 | | Immediate-early 1 protein (IE1) of rhesus macaque cytomegalovirus | | Descriptor: | RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Controlled crystal dehydration triggers a space-group switch and shapes the tertiary structure of cytomegalovirus immediate-early 1 (IE1) protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9H4U
 
 | |
7P7U
 
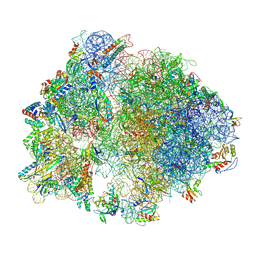 | | E. faecalis 70S ribosome with P-tRNA, state IV | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
6VUP
 
 | |
7OU8
 
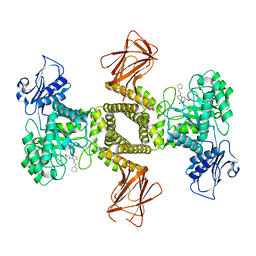 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7PC4
 
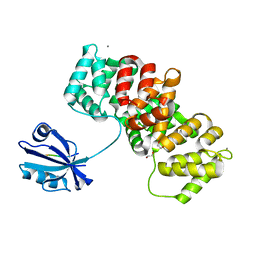 | | The PDZ domain of SNTB1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-1-syntrophin,Annexin A2, CALCIUM ION, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6OS2
 
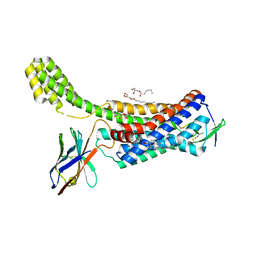 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV026 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
4RJ8
 
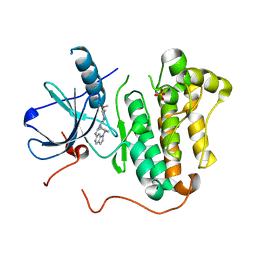 | | EGFR kinase (T790M/L858R) with inhibitor compound 8 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-pyrrolo[3,2-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ7
 
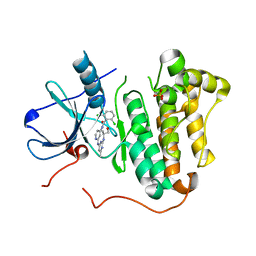 | | EGFR kinase (T790M/L858R) with inhibitor compound 1 | | Descriptor: | 2,6-dichloro-N-{2-[(2-{[(2S)-1-hydroxypropan-2-yl]amino}-6-methylpyrimidin-4-yl)amino]pyridin-4-yl}benzamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
8AI6
 
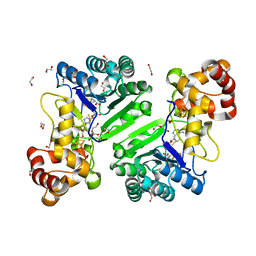 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
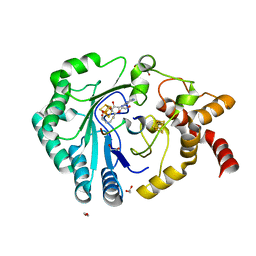 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
