3PDF
 
 | | Discovery of Novel Cyanamide-Based Inhibitors of Cathepsin C | | Descriptor: | 2,5-dibromo-N-{(3R,5S)-1-[(Z)-iminomethyl]-5-methylpyrrolidin-3-yl}benzenesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, B, Laine, D. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of novel cyanamide-based inhibitors of cathepsin C.
Acs Med.Chem.Lett., 2, 2011
|
|
4D4X
 
 | | Nitrosyl complex of the D121I variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
3CIZ
 
 | |
2L45
 
 | | C-terminal zinc knuckle of the HIVNCp7 with DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
1HZK
 
 | |
4M6X
 
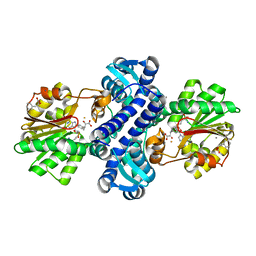 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LPH
 
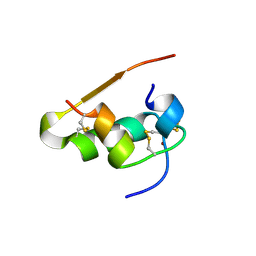 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
4M72
 
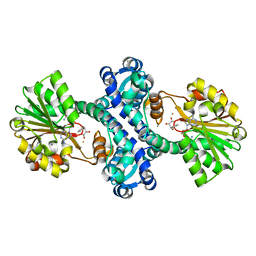 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2R,3R)-2-hydroxy-3-methoxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NNG
 
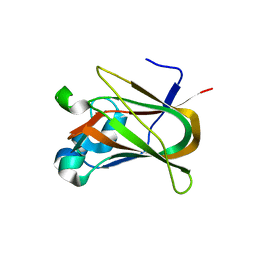 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | Descriptor: | CALCIUM ION, uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
2ZC7
 
 | |
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
4M74
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2S,3R)-2,3-dihydroxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
2XM4
 
 | |
3L48
 
 | |
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
4QL1
 
 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
3I5N
 
 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4J93
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
4MJN
 
 | |
2XM0
 
 | |
2XLW
 
 | |
1QN2
 
 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
2JNH
 
 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
3QJH
 
 | | The crystal structure of the 5c.c7 TCR | | Descriptor: | 5c.c7 alpha chain, 5c.c7 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
