8VK4
 
 | |
8VK3
 
 | |
8VK2
 
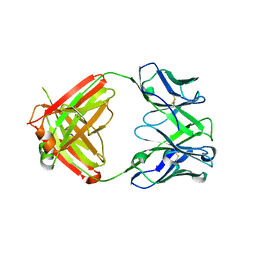 | | X-ray crystal structure of human IgE 4C8 Fab | | Descriptor: | IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 2024
|
|
8VK1
 
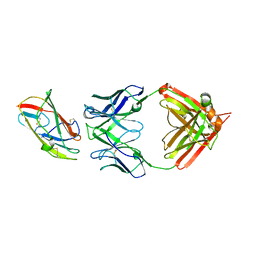 | | X-ray crystal structure of human IgE 4C8 Fab complex with Der p 2.0103 | | Descriptor: | Der p 2 variant 3, IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 2024
|
|
8VJS
 
 | |
8VJR
 
 | |
8VJP
 
 | | Histidine-covalent stapled alpha-helical peptide (155H1) targeting hMcl-1 | | Descriptor: | (4Z)-oct-4-en-1-ol, (S~1~R)-3-carbamoyl-4-methoxybenzene-1-sulfinic acid, Histidine-covalent stapled alpha-helical peptide, ... | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-01-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Histidine-Covalent Stapled Alpha-Helical Peptides Targeting hMcl-1.
J.Med.Chem., 67, 2024
|
|
8VJO
 
 | |
8VJN
 
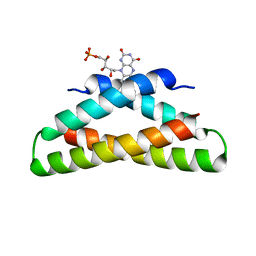 | |
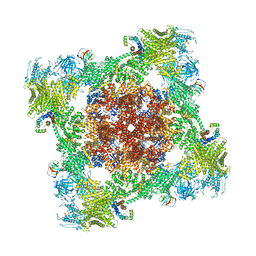
8VJK
 
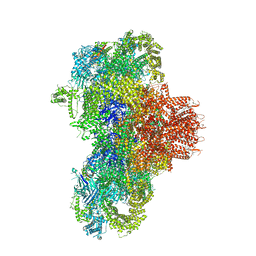 | | Structure of mouse RyR1 (high-Ca2+/CFF/ATP dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VJJ
 
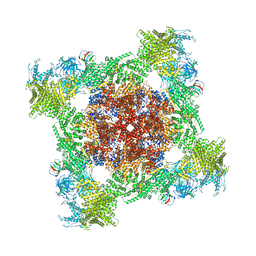 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
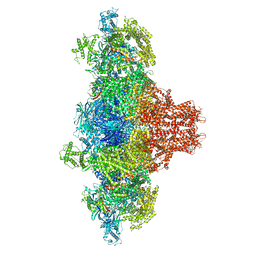
8VJC
 
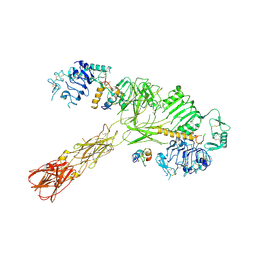 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJB
 
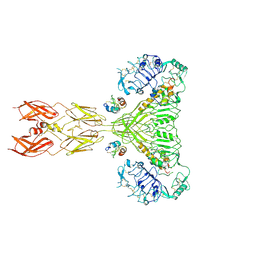 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJ7
 
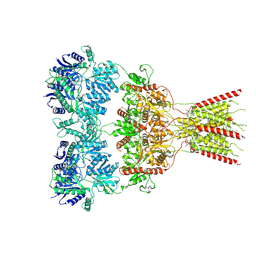 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
8VJ6
 
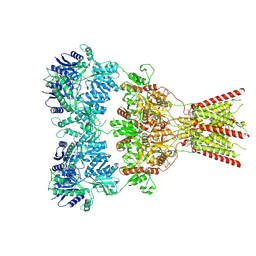 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
8VJ3
 
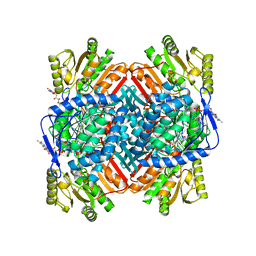 | |
8VJ2
 
 | |
8VJ1
 
 | |
8VIS
 
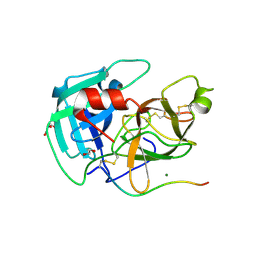 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide
To Be Published
|
|
8VIJ
 
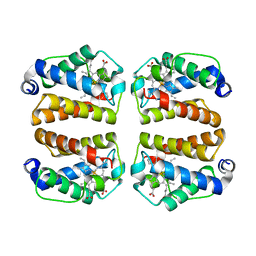 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant (cyanomet) | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lecomte, J.T.J, Schlessman, J.L, Martinez, J.E, Schultz, T.D, Siegler, M.A, DelCampo, M, Le Magueres, P. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant in the cyanomet state
To be published
|
|
8VID
 
 | |
8VIB
 
 | |
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
