9FB4
 
 | |
9FA1
 
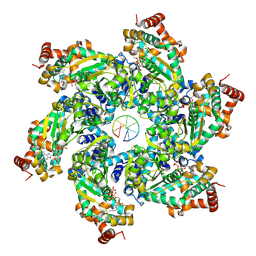 | |
9F9X
 
 | |
7R0A
 
 | | Structure of sarin phosphonylated acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7R7B
 
 | |
2WLA
 
 | | Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, GLYCEROL | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
6N96
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2~{R})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, (2~{S})-1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethoxy]-1-oxidanylidene-propane-2-sulfonic acid, IMIDAZOLE, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7R02
 
 | | Mus musculus acetylcholinesterase in complex with N-(3-(diethylamino)propyl)-4-methyl-3-nitrobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
9F39
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-54 | | Descriptor: | (2R,3R)-3-[[(2S)-3-cyclopropyl-2-[3-(2-methylpropanoylamino)-2-oxidanylidene-pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3OOI
 
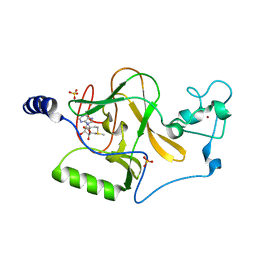 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
9FJX
 
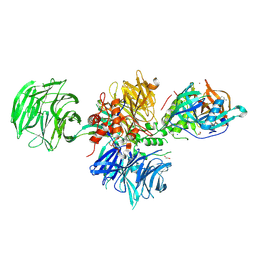 | |
7RM6
 
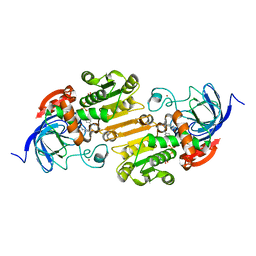 | |
7QVZ
 
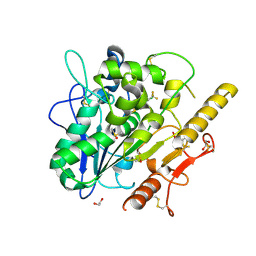 | | ARUK3001043_Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Hillier, J, Willis, N.J, Mahy, W, Fish, P, Jones, E.Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure Activity Analysis of Notum Fragment Screen Hits with Development
To Be Published
|
|
9EKE
 
 | |
9F73
 
 | |
7DA7
 
 | |
174L
 
 | |
9FHQ
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR04 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-6-oxidanylidene-pyrimidin-5-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9EKD
 
 | |
7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | Descriptor: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DAS
 
 | |
