9B0Z
 
 | |
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
9C5S
 
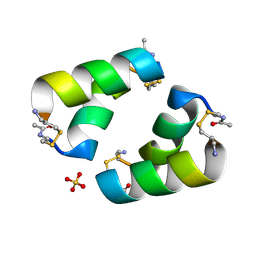 | | Disulfide-linked, antiparallel p53-derived peptide dimer (CV1) | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Vithanage, N, Kreitler, D.K, DiGiorno, M.C, Victorio, C.G, Sawyer, N, Outlaw, V.K. | | Deposit date: | 2024-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
Res Sq, 2024
|
|
8Y6F
 
 | |
8X5Y
 
 | |
9B20
 
 | |
8VEC
 
 | | Deep Mutational Scanning of SARS-CoV-2 PLpro | | Descriptor: | Papain-like protease nsp3, ZINC ION | | Authors: | Wu, X, Nguyen, J.V, Call, M.E, Call, M.J. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational profiling of SARS-CoV-2 papain-like protease reveals requirements for function, structure, and drug escape.
Nat Commun, 15, 2024
|
|
8XHK
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
9ASZ
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenylethyl 2-pyrrolidone inhibitor | | Descriptor: | (1S,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8XT1
 
 | | Cryo-EM structure of the human 39S mitoribosome with 5uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8W8T
 
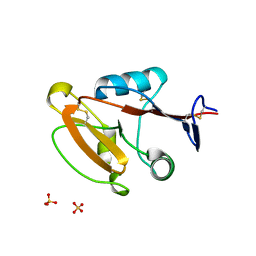 | | Crystal structure of human CLEC12A CRD | | Descriptor: | C-type lectin domain family 12 member A, SULFATE ION | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
9C0C
 
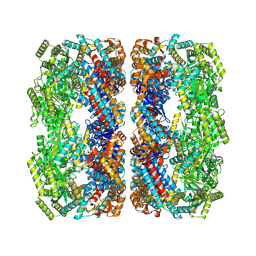 | | E.coli GroEL apoenzyme | | Descriptor: | 60 kDa chaperonin | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
8WOO
 
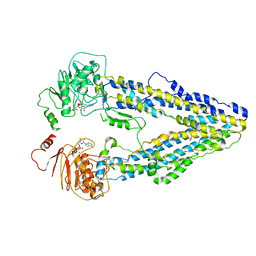 | | Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state | | Descriptor: | ABC transporter B family member 19, Brassinolide, MAGNESIUM ION, ... | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8XIF
 
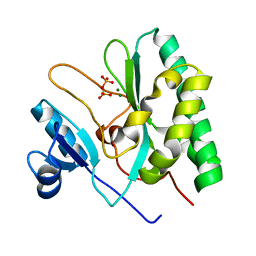 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8VSA
 
 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
8XIM
 
 | |
9EWM
 
 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8XDO
 
 | |
8XPT
 
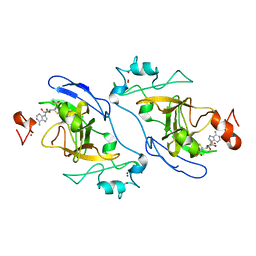 | | The Crystal Structure of EHMT1 from Biortus. | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2024-01-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Crystal Structure of EHMT1 from Biortus.
To Be Published
|
|
9EOW
 
 | |
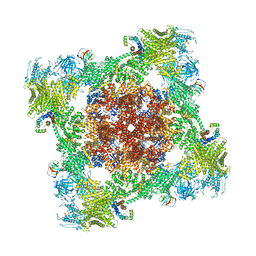
8VK4
 
 | |
9EQF
 
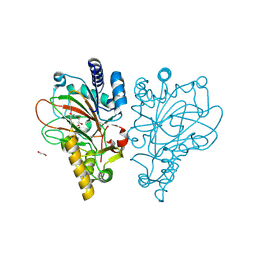 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
8XVC
 
 | | CryoEM structure of ADP-DNA-MuB conformation1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
9FX6
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8VRM
 
 | | Crystal structure of the Pcryo_0619 N-acetyltransferase from Psychrobacter cryohalolentis K5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Acetyltransferase, ... | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-22 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
