2ET8
 
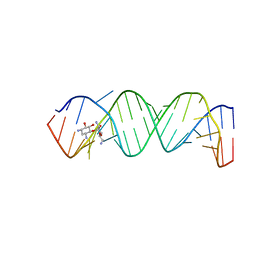 | | Complex Between Neamine and the 16S-RRNA A-Site | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
1V3L
 
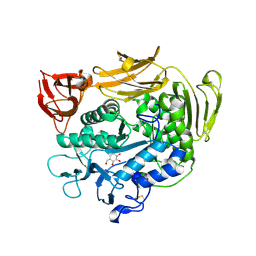 | | Crystal structure of F283L mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
1ZLY
 
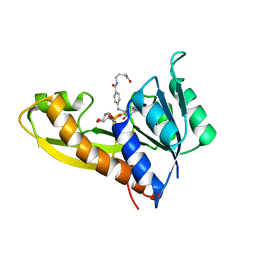 | | The structure of human glycinamide ribonucleotide transformylase in complex with alpha,beta-N-(hydroxyacetyl)-D-ribofuranosylamine and 10-formyl-5,8,dideazafolate | | Descriptor: | 4-[(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)METHYL]AMINO}BENZOYL)AMINO]BUTANOIC ACID, 5-O-phosphono-beta-D-ribofuranosylamine, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E.S, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
1ZGS
 
 | |
3U65
 
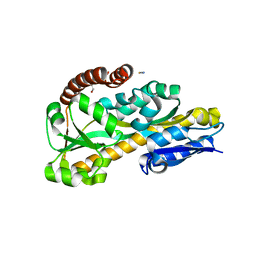 | | The Crystal Structure of Tat-P(T) (Tp0957) | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, Tp33 protein | | Authors: | Brautigam, C.A, Tomchick, D.R, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1ZLR
 
 | | Factor XI catalytic domain complexed with 2-guanidino-1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethyl nicotinate | | Descriptor: | (1R)-2-{[AMINO(IMINO)METHYL]AMINO}-1-{4-[(4R)-4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL]PHENYL}ETHYL NICOTINATE, Coagulation factor XI, GLYCEROL, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R, Strickler, J. | | Deposit date: | 2005-05-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2D4Q
 
 | | Crystal structure of the Sec-PH domain of the human neurofibromatosis type 1 protein | | Descriptor: | Neurofibromin, OXTOXYNOL-10, PYROPHOSPHATE 2- | | Authors: | D'angelo, I, Welti, S, Bonneau, F, Scheffzek, K. | | Deposit date: | 2005-10-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein
Embo Rep., 7, 2006
|
|
2FFR
 
 | | Crystallographic studies on N-azido-beta-D-glucopyranosylamine, an inhibitor of glycogen phosphorylase: comparison with N-acetyl-beta-D-glucopyranosylamine | | Descriptor: | Glycogen phosphorylase, muscle form, N-(azidoacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Petsalakis, E.I, Chrysina, E.D, Tiraidis, C, Hadjiloi, T, Leonidas, D.D, Oikonomakos, N.G, Aich, U, Varghese, B, Loganathan, D. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic studies on N-azidoacetyl-beta-d-glucopyranosylamine, an inhibitor of glycogen phosphorylase: Comparison with N-acetyl-beta-d-glucopyranosylamine.
Bioorg.Med.Chem., 14, 2006
|
|
2F6D
 
 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
3PWY
 
 | | Crystal structure of an extender (SPD28345)-modified human PDK1 complex 2 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, N-[2-({6-[(2-sulfanylethyl)amino]pyrimidin-4-yl}amino)ethyl]propanamide | | Authors: | Elling, R.A, Penny, D.M, Simmons, R.L, Erlanson, D.A, Romanowski, M.J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2DTW
 
 | | Crystal Structure of basic winged bean lectin in complex with 2Me-O-D-Galactose | | Descriptor: | 2-O-methyl-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-07-18 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the carbohydrate-specificity of basic winged-bean lectin and its differential affinity for Gal and GalNAc
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1LMA
 
 | |
1Z2N
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase complexed Mg2+/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, inositol 1,3,4-trisphosphate 5/6-kinase | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
2DKO
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | Caspase-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Mittl, P.R.E, Ganesan, R, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
2ZOZ
 
 | | Crystal structure of the ethidium-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | ETHIDIUM, GLYCEROL, SULFATE ION, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2DU0
 
 | | Crystal structure of basic winged bean lectin in complex with Alpha-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-07-19 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the carbohydrate-specificity of basic winged-bean lectin and its differential affinity for Gal and GalNAc
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DCN
 
 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
3UC8
 
 | | Trp-cage cyclo-TC1 - tetragonal crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, cyclo-TC1 | | Authors: | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2D1J
 
 | | Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide | | Descriptor: | 2-({4-[(5-CHLORO-1H-INDOL-2-YL)SULFONYL]PIPERAZIN-1-YL}CARBONYL)THIENO[3,2-B]PYRIDINE 4-OXIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological activity of non-basic compounds as factor Xa inhibitors: SAR study of S1 and aryl binding sites.
Bioorg.Med.Chem., 13, 2005
|
|
3B4N
 
 | | Crystal Structure Analysis of Pectate Lyase PelI from Erwinia chrysanthemi | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Endo-pectate lyase, ... | | Authors: | Creze, C, Castang, S, Derivery, E, Haser, R, Shevchik, V, Gouet, P. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia chrysanthemi in Complex with Its Substrate.
J.Biol.Chem., 283, 2008
|
|
3OS2
 
 | | PFV target capture complex (TCC) at 3.32 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The mechanism of retroviral integration from X-ray structures of its key intermediates
Nature, 468, 2010
|
|
2F4S
 
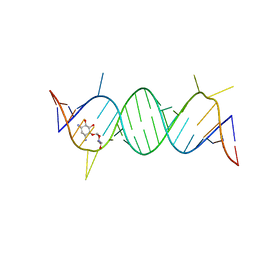 | | A-site RNA in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2ESU
 
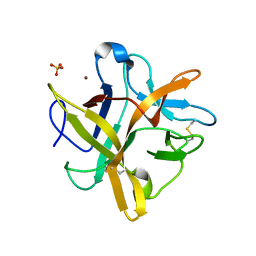 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
