4B1H
 
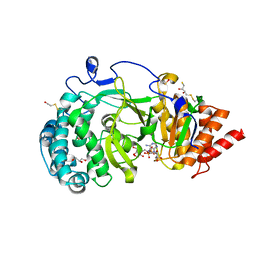 | | Structure of human PARG catalytic domain in complex with ADP-ribose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
1OMD
 
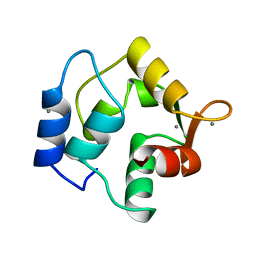 | | STRUCTURE OF ONCOMODULIN REFINED AT 1.85 ANGSTROMS RESOLUTION. AN EXAMPLE OF EXTENSIVE MOLECULAR AGGREGATION VIA CA2+ | | Descriptor: | CALCIUM ION, ONCOMODULIN | | Authors: | Ahmed, F.R, Przybylska, M, Rose, D.R, Birnbaum, G.I, Pippy, M.E, Macmanus, J.P. | | Deposit date: | 1990-04-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oncomodulin refined at 1.85 A resolution. An example of extensive molecular aggregation via Ca2+.
J.Mol.Biol., 216, 1990
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1XPR
 
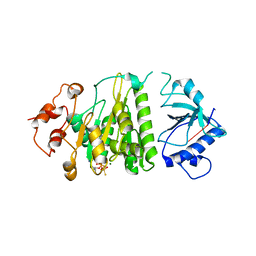 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-formylbicyclomycin (FB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-FORMYLBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1PDO
 
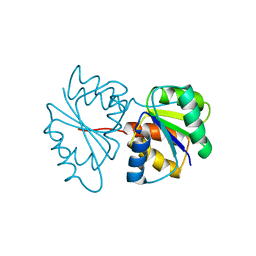 | |
4C8Q
 
 | | Crystal structure of the yeast Lsm1-7-Pat1 complex | | Descriptor: | COBALT (II) ION, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, SM-LIKE PROTEIN LSM1, ... | | Authors: | Sharif, H, Conti, E. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic Mrna Turnover
Cell Rep., 5, 2013
|
|
1PFR
 
 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (III) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-12-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
4COI
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with glycerol in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, GLYCEROL, ZINC ION | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4CON
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with citrate in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CITRIC ACID | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COM
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COL
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with dATP bound in the specificity site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, MAGNESIUM ION, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
1Q14
 
 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | Descriptor: | CHLORIDE ION, HST2 protein, ZINC ION | | Authors: | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
4B1J
 
 | | Structure of human PARG catalytic domain in complex with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Overman, R, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4B1I
 
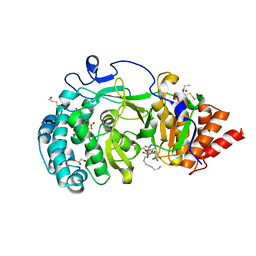 | | Structure of human PARG catalytic domain in complex with OA-ADP-HPD | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 8-n-octylamino-adenosine diphosphate hydroxypyrrolidinediol, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, Johnson, T, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
3K1L
 
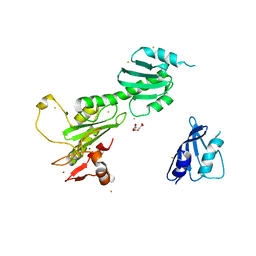 | | Crystal Structure of FANCL | | Descriptor: | CITRIC ACID, Fancl, GOLD ION, ... | | Authors: | Cole, A.R, Walden, H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the catalytic subunit FANCL of the Fanconi anemia core complex
Nat.Struct.Mol.Biol., 17, 2010
|
|
1XPU
 
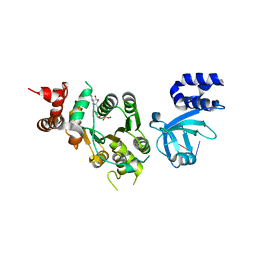 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1O15
 
 | |
1NLR
 
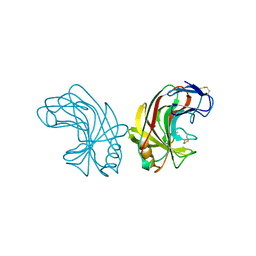 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
1XPO
 
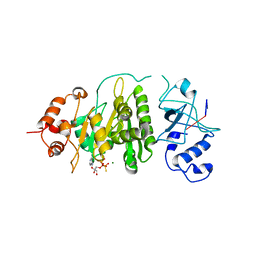 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
4BRW
 
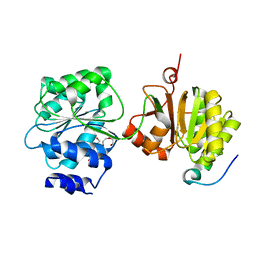 | | Crystal structure of the yeast Dhh1-Pat1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-DEPENDENT RNA HELICASE DHH1, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, ... | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
1QL2
 
 | |
3KN1
 
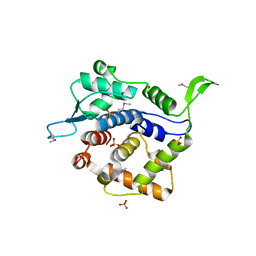 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | Descriptor: | Golgi phosphoprotein 3, SULFATE ION | | Authors: | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
2A8V
 
 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
4COJ
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima in complex with dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
1J23
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
