1Q3Q
 
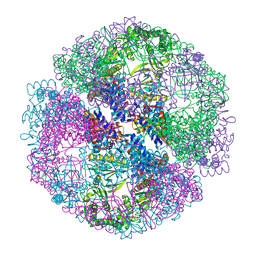 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (two-point mutant complexed with AMP-PNP) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
5KY6
 
 | |
1PNT
 
 | |
4BCQ
 
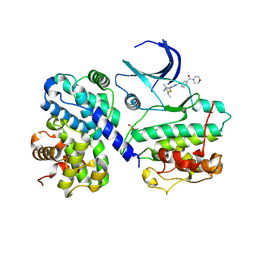 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]-2-{[3-(morpholin-4-ylcarbonyl)phenyl]amino}pyrimidine-5-carbonitrile, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
2I5T
 
 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | Descriptor: | Protein C7orf24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
1CDD
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | PHOSPHATE ION, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1Q3S
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1BQ2
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
4KES
 
 | | Crystal structure of SsoPox W263T | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
4KFD
 
 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 6.0 | | Descriptor: | COPPER (II) ION, GLYCEROL, HYDROGEN PEROXIDE, ... | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
1PNG
 
 | |
8THJ
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
1RAP
 
 | |
6AVT
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.5 WITH CROSS-LINKING TO FIRST BASE TEMPLATE OVERHANG | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-09-04 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
1R6E
 
 | |
5KTR
 
 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound maleate and Fe4S4 cluster | | Descriptor: | AMMONIUM ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
3TWX
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human FNBP1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3PKQ
 
 | | Q83D Variant of S. Enterica RmlA with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
5KTP
 
 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound itaconate and Fe4S4 cluster | | Descriptor: | 2-methylidenebutanedioic acid, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
3TL9
 
 | | crystal structure of HIV protease model precursor/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3TNE
 
 | | The crystal structure of protease Sapp1p from Candida parapsilosis in complex with the HIV protease inhibitor ritonavir | | Descriptor: | RITONAVIR, Secreted aspartic protease | | Authors: | Dostal, J, Brynda, J, Hruskova-Heidingsfeldova, O, Pachl, P, Pichova, I, Rezacova, P. | | Deposit date: | 2011-09-01 | | Release date: | 2012-03-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of protease Sapp1p from Candida parapsilosis in complex with the HIV protease inhibitor ritonavir.
J Enzyme Inhib Med Chem, 27, 2012
|
|
5KTT
 
 | |
1RIN
 
 | | X-RAY CRYSTAL STRUCTURE OF A PEA LECTIN-TRIMANNOSIDE COMPLEX AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEA LECTIN, ... | | Authors: | Rini, J.M, Hardman, K.D, Einspahr, H, Suddath, F.L, Carver, J.P. | | Deposit date: | 1993-01-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of a pea lectin-trimannoside complex at 2.6 A resolution.
J.Biol.Chem., 268, 1993
|
|
