2DR0
 
 | |
1TWB
 
 | | SspB disulfide crosslinked to an ssrA degradation tag | | Descriptor: | Stringent starvation protein B homolog, ssrA peptide | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
1U04
 
 | |
2DUD
 
 | |
1U1R
 
 | |
1U1L
 
 | |
1U2O
 
 | | Crystal Structure Of The N-Domain Of Grp94 Lacking The Charged Domain In Complex With Neca | | Descriptor: | Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE, PENTAETHYLENE GLYCOL, ... | | Authors: | Soldano, K.L, Jivan, A, Nicchitta, C.V, Gewirth, D.T. | | Deposit date: | 2004-07-19 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation
J.Biol.Chem., 278, 2003
|
|
1VNE
 
 | |
2DQH
 
 | | Crystal structure of hyhel-10 FV mutant (Hy58a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
1VKJ
 
 | | Crystal structure of heparan sulfate 3-O-sulfotransferase isoform 1 in the presence of PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULFATE ION, heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | | Authors: | Thorp, S, Lee, K.A, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1
J.Biol.Chem., 279, 2004
|
|
2DQV
 
 | | Structure of the C-terminal lobe of bovine lactoferrin in complex with galactose at 2.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-05-31 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the C-terminal lobe of bovine lactoferrin in complex with galactose at 2.7 A resolution
To be Published
|
|
1VNF
 
 | |
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | DrP35 | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
1VPN
 
 | | UNASSEMBLED POLYOMAVIRUS VP1 PENTAMER | | Descriptor: | POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
1VPT
 
 | | AS11 VARIANT OF VACCINIA VIRUS PROTEIN VP39 IN COMPLEX WITH S-ADENOSYL-L-METHIONINE | | Descriptor: | S-ADENOSYLMETHIONINE, VP39 | | Authors: | Hodel, A.E, Gershon, P.D, Shi, X, Quiocho, F.A. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends.
Cell(Cambridge,Mass.), 85, 1996
|
|
2DSZ
 
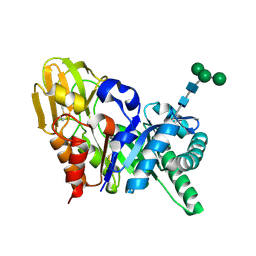 | | Three dimensional structure of a goat signalling protein secreted during involution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Singh, N, Ujwal, R, Srivastava, D.B, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1W02
 
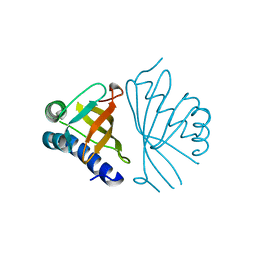 | |
1RAF
 
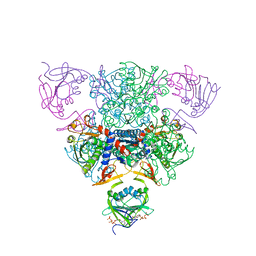 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RBI
 
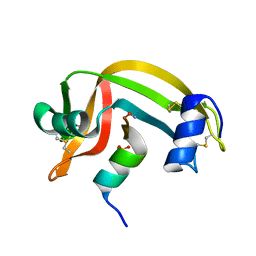 | |
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W14
 
 | | UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | N-[(2-PHENYLETHYL)SULFONYL]-D-SERYL-N-[(1S)-4-[(DIAMINOMETHYLENE)AMINO]-1-(HYDROXYMETHYL)BUTYL]-L-ALANINAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystals of Urokinase Type Plasminogen Activator Complexes Reveal the Binding Mode of Peptidomimetic Inhibitors.
J.Mol.Biol., 328, 2003
|
|
1W1A
 
 | | Structure of Bacillus subtilis PdaA in complex with NAG, a family 4 Carbohydrate esterase. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Blair, D.E, van Aalten, D.M.F. | | Deposit date: | 2004-06-18 | | Release date: | 2005-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine.
FEBS Lett., 570, 2004
|
|
1RKR
 
 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BG6
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
1RLI
 
 | |
