8CRJ
 
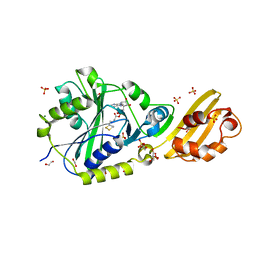 | | Crystal structure of LplA1 in complex with lipoyl-AMP (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5FM4
 
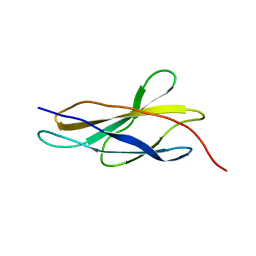 | |
1KT9
 
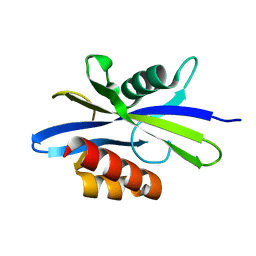 | | Crystal Structure of C. elegans Ap4A Hydrolase | | Descriptor: | Diadenosine Tetraphosphate Hydrolase | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
4MSO
 
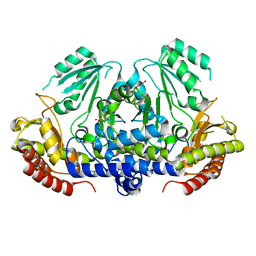 | |
7AGR
 
 | |
8CQG
 
 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis | | Descriptor: | 1,2-ETHANEDIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Skagseth, S, Lund, B.A, Griese, J.J, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
1KOU
 
 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6T6K
 
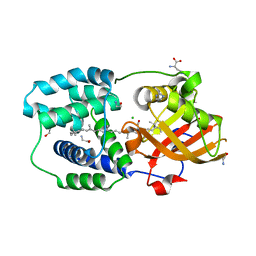 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T6W
 
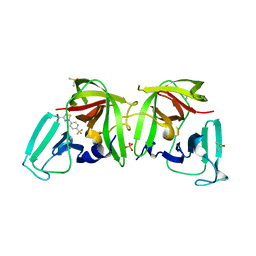 | |
7AGP
 
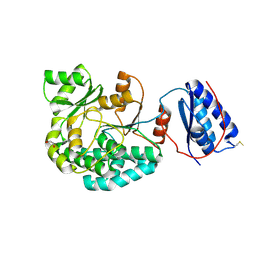 | | Structure of the AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Mycocerosic acid synthase | | Authors: | Brison, Y, Nahoum, V, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
8COL
 
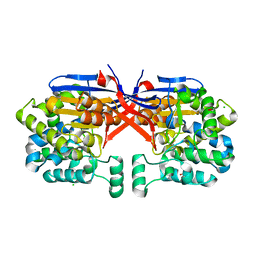 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (orthorombic form R4oP-2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imioloczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CHI
 
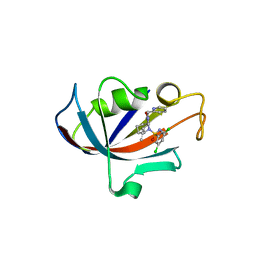 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
4MTR
 
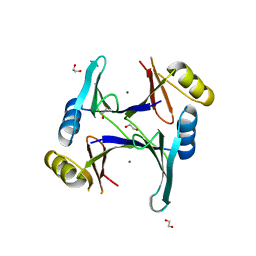 | | Zn-bound GloA2 | | Descriptor: | 1,2-ETHANEDIOL, Lactoylglutathione lyase, ZINC ION | | Authors: | Bythell-Douglas, R, Bond, C.S. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of a homodimeric Pseudomonas glyoxalase I enzyme reveals asymmetric metallation commensurate with half-of-sites activity.
Chemistry, 21, 2015
|
|
2YPV
 
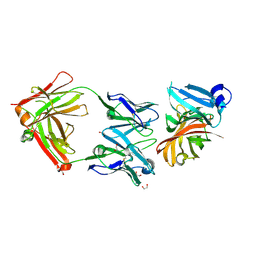 | | Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody | | Descriptor: | 1,2-ETHANEDIOL, FAB 12C1, LIPOPROTEIN | | Authors: | Malito, E, Veggi, D, Bottomley, M.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining a Protective Epitope on Factor H Binding Protein, a Key Meningococcal Virulence Factor and Vaccine Antigen.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6THS
 
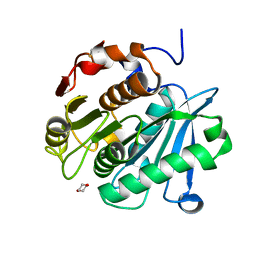 | |
6T3D
 
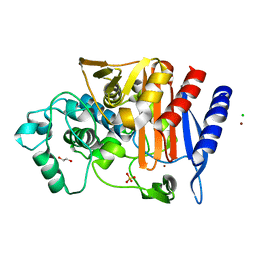 | | Crystal structure of AmpC from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
4NA9
 
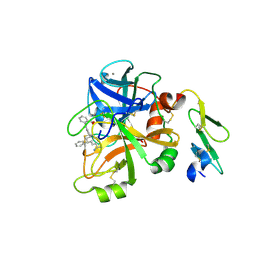 | | Factor VIIa in complex with the inhibitor 3'-amino-5'-[(2s,4r)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid | | Descriptor: | 3'-amino-5'-[(2S,4R)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
4NAE
 
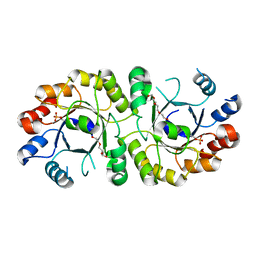 | | PcrB from Geobacillus kaustophilus, with bound G1P | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Heptaprenylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Peterhoff, D, Beer, B, Rajendran, C, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-10-22 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
1KVR
 
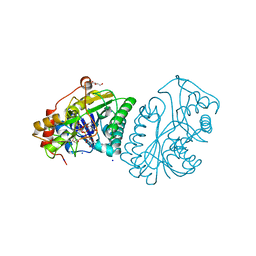 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
2YGW
 
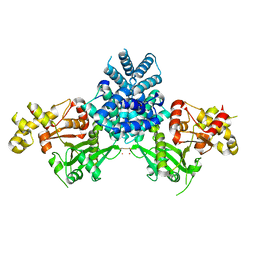 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
6DS9
 
 | | Elongated version of a de novo designed three helix bundle structure (GRa3D) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Koebke, K.J, Ruckthong, L.R, Meagher, J.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Clarifying the Copper Coordination Environment in a de Novo Designed Red Copper Protein.
Inorg Chem, 57, 2018
|
|
4MXC
 
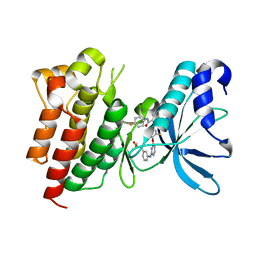 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[2-({3-[(methylsulfonyl)methyl]phenyl}amino)pyrimidin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Discovery of Anilinopyrimidines as Dual Inhibitors of c-Met and VEGFR-2: Synthesis, SAR, and Cellular Activity
ACS MED.CHEM.LETT., 5, 2014
|
|
5FNS
 
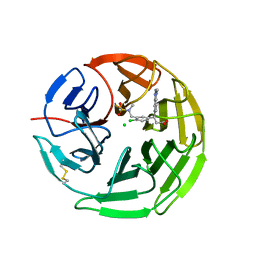 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2YQD
 
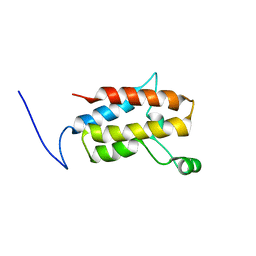 | |
