8OPC
 
 | |
3KQS
 
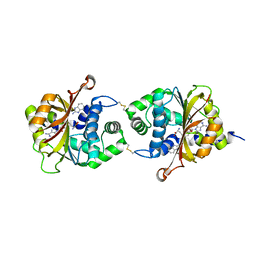 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Aminobenzimidazole | | Descriptor: | 1H-benzimidazol-2-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
4OC5
 
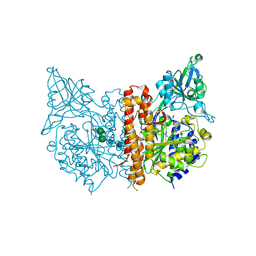 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CHIBzL, a urea-based inhibitor N~2~-{[(S)-carboxy(4-hydroxyphenyl)methyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5VS4
 
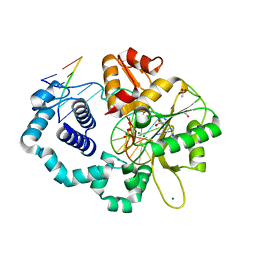 | | Human DNA polymerase beta 8-oxoG:dA extension with dTTP after 120 s | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*(8OG)P*GP*CP*GP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*TP*GP*CP*GP*CP*AP*T)-3'), ... | | Authors: | Reed, A.J, Suo, Z. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Time-Dependent Extension from an 8-Oxoguanine Lesion by Human DNA Polymerase Beta.
J. Am. Chem. Soc., 139, 2017
|
|
5VTI
 
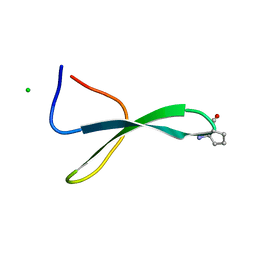 | | Structure of Pin1 WW Domain Sequence 3 with [R,R]-ACPC Loop Substitution | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Kreitler, D.F, Thomas, N.C, Gellman, S.H, Forest, K.T. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of beta-Amino Acid Replacements in Protein Loops: Effects on Conformational Stability and Structure.
Chembiochem, 19, 2018
|
|
8OPD
 
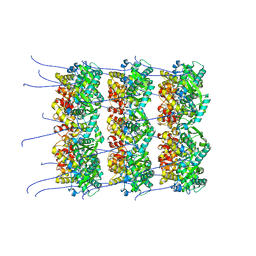 | |
2G09
 
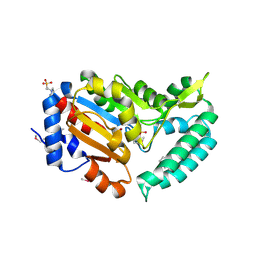 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
7YMS
 
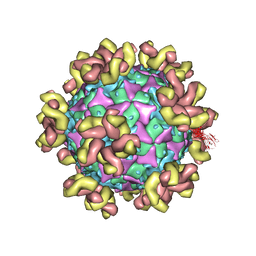 | |
5VXX
 
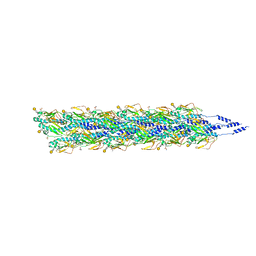 | | Cryo-EM reconstruction of Neisseria gonorrhoeae Type IV pilus | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose | | Authors: | Wang, F, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
4OQE
 
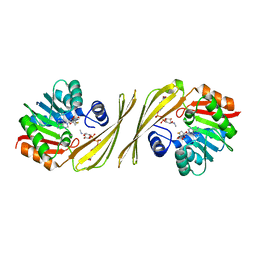 | | Crystal structure of the tylM1 N,N-dimethyltransferase in complex with SAH and TDP-Fuc3NMe | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-N-methylamino-3,6-dideoxygalactose, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Production of a novel N-monomethylated dideoxysugar.
Biochemistry, 53, 2014
|
|
5VXY
 
 | | Cryo-EM reconstruction of PAK pilus from Pseudomonas aeruginosa | | Descriptor: | Fimbrial protein | | Authors: | Wang, F, Osinksi, T, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
5W3O
 
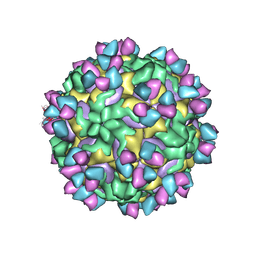 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8CHS
 
 | |
3KZU
 
 | |
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
3L3X
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
2G57
 
 | | Structure of the Phosphorylation Motif of the oncogenic Protein beta-Catenin Recognized By a Selective Monoclonal Antibody | | Descriptor: | Beta-catenin | | Authors: | Megy, S, Bertho, G, Gharbi-Benarous, J, Baleux, F, Benarous, R, Girault, J.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | STD and TRNOESY NMR studies for the epitope mapping of the phosphorylation motif of the oncogenic protein beta-catenin recognized by a selective monoclonal antibody
Febs Lett., 580, 2006
|
|
8C50
 
 | |
3L4G
 
 | | Crystal structure of Homo Sapiens cytoplasmic Phenylalanyl-tRNA synthetase | | Descriptor: | PHENYLALANINE, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Finarov, I, Moor, N, Kessler, N, Klipcan, L, Safro, M.G. | | Deposit date: | 2009-12-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
Structure, 18, 2010
|
|
2G0G
 
 | | Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities | | Descriptor: | 3-FLUORO-N-[1-(4-FLUOROPHENYL)-3-(2-THIENYL)-1H-PYRAZOL-5-YL]BENZENESULFONAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Huang, C.F, Lin, Y.T, Hsu, J.T.A, Wu, S.Y. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure-Based Drug Design of a Novel Family of PPARgamma Partial Agonists: Virtual Screening, X-ray Crystallography, and in Vitro/in Vivo Biological Activities
J.Med.Chem., 49, 2006
|
|
4O7E
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-610251-B | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-phenyl-4-(pyridin-4-yl)-1H-imidazol-5-yl]phenol, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
3KQL
 
 | |
3KPV
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Adenine | | Descriptor: | ADENINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
5W68
 
 | | Type II secretin from Enteropathogenic Escherichia coli - GspD | | Descriptor: | Putative type II secretion protein | | Authors: | Hay, I.D, Belousoff, M.J, Dunstan, R, Bamert, R, Lithgow, T. | | Deposit date: | 2017-06-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and Membrane Topography of the Vibrio-Type Secretin Complex from the Type 2 Secretion System of Enteropathogenic Escherichia coli.
J. Bacteriol., 200, 2018
|
|
3KQY
 
 | |
