3BLR
 
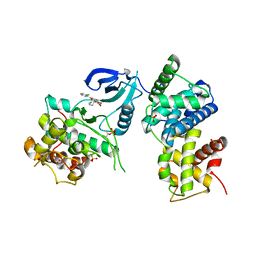 | | Crystal Structure of Human CDK9/cyclinT1 in complex with Flavopiridol | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, Cell division protein kinase 9, Cyclin-T1, ... | | Authors: | Baumli, S, Lolli, G, Lowe, E.D, Johnson, L.N. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation
Embo J., 27, 2008
|
|
3TN8
 
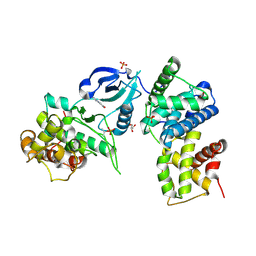 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.E. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3TNH
 
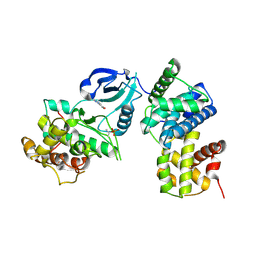 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3TNI
 
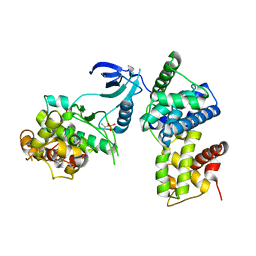 | | structure of CDK9/cyclin T F241L | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.234 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
4I9F
 
 | | Crystal structure of glycerol phosphate phosphatase Rv1692 from Mycobacterium tuberculosis in complex with calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycerol 3-phosphate phosphatase | | Authors: | Biswas, T, Larrouy-Maumus, G, de Carvalho, L.P, Tsodikov, O.V. | | Deposit date: | 2012-12-05 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BCG
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
4BCJ
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[(3-hydroxyphenyl)amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4BCF
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(4-ethanoyl-1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4BCH
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 4-(4-methyl-2-methylimino-3H-1,3-thiazol-5-yl)-2-[(4-methyl-3-morpholin-4-ylsulfonyl-phenyl)amino]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4BCI
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 3-[[5-cyano-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidin-2-yl]amino]benzenesulfonamide, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4EC9
 
 | |
4EC8
 
 | |
4I9G
 
 | | Crystal structure of glycerol phosphate phosphatase Rv1692 from Mycobacterium tuberculosis in complex with magnesium | | Descriptor: | Glycerol 3-phosphate phosphatase, MAGNESIUM ION | | Authors: | Biswas, T, Larrouy-Maumus, G, de Carvalho, L.P, Tsodikov, O.V. | | Deposit date: | 2012-12-05 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1MJ9
 
 | | Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A | | Descriptor: | COENZYME A, ESA1 PROTEIN, SODIUM ION | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJB
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJA
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase domain complexed with acetyl coenzyme A | | Descriptor: | COENZYME A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
6ERD
 
 | | Crystal structure of a putative acetyltransferase from Bacillus cereus species. | | Descriptor: | Aminoglycoside N6'-acetyltransferase, CHLORIDE ION, GLYCEROL | | Authors: | Silvestre, H.L, Bolanos-Garcia, V.M, Asensio, J.L, Blundell, T.L, Bastida, A. | | Deposit date: | 2017-10-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterisation of RimL from Bacillus cereus, a new N alpha-acetyltransferase of ribosomal proteins that was wrongly assigned as an aminoglycosyltransferase.
Int.J.Biol.Macromol., 263, 2024
|
|
4W4U
 
 | |
4U5Y
 
 | | Crystal Structure of the complex between the GNAT domain of S. lividans PAT and the acetyl-CoA synthetase C-terminal domain of S. enterica | | Descriptor: | Acetyl-coenzyme A synthetase, Acyl-CoA synthetase, SULFATE ION | | Authors: | Taylor, K.C, Tucker, A.C, Rank, K.C, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2014-07-25 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Insights into the specificity of lysine acetyltransferases.
J.Biol.Chem., 289, 2014
|
|
3SMA
 
 | |
2VEZ
 
 | | AfGNA1 crystal structure complexed with Acetyl-CoA and Glucose-6P gives new insights into catalysis | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, PHOSPHATE ION, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O, Shepherd, S, van Aalten, D.M.F. | | Deposit date: | 2007-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glucose-6-Phosphate as a Probe for the Glucosamine- 6-Phosphate N-Acetyltransferase Michaelis Complex.
FEBS Lett., 581, 2007
|
|
7Q3A
 
 | | Crystal structure of MAB_4324 a tandem repeat GNAT from Mycobacterium abscessus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2021-10-27 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical, structural, and functional studies reveal that MAB_4324c from Mycobacterium abscessus is an active tandem repeat N-acetyltransferase.
Febs Lett., 596, 2022
|
|
7TIF
 
 | |
4WHN
 
 | | Structure of toxin-activating acyltransferase (TAAT) | | Descriptor: | ApxC, CITRIC ACID | | Authors: | Crow, A, Greene, N.P, Hughes, C, Koronakis, V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a bacterial toxin-activating acyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3LQ5
 
 | | Structure of CDK9/CyclinT in complex with S-CR8 | | Descriptor: | (2S)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Hole, A.J, Endicott, J.A, Baumli, S. | | Deposit date: | 2010-02-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CDK Inhibitors Roscovitine and CR8 Trigger Mcl-1 Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells
Genes Cancer, 1, 2010
|
|
