3GY9
 
 | |
3GZ7
 
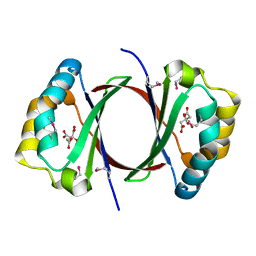 | |
3GZE
 
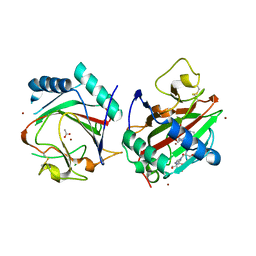 | |
3GZ4
 
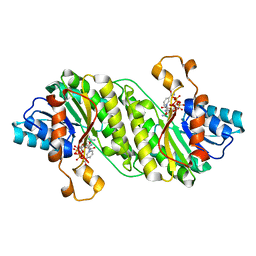 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3GZI
 
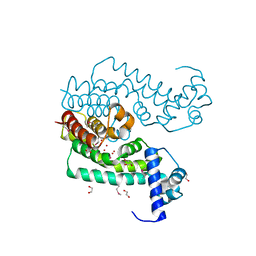 | |
3H12
 
 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
3H0N
 
 | |
3IHI
 
 | | Crystal structure of mouse thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2009-07-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
3IK5
 
 | |
3ILL
 
 | | Crystal structure of E. coli HPPK(D97A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
3IJY
 
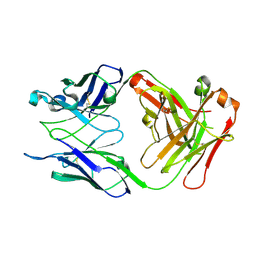 | | Structure of S67-27 in Complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3ILT
 
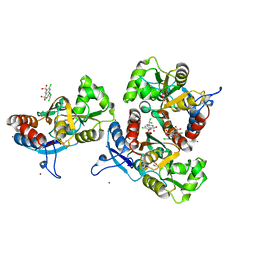 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | Descriptor: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IMW
 
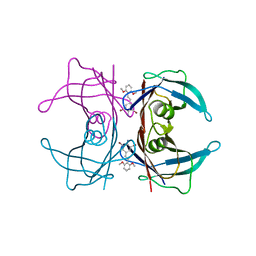 | | Transthyretin in complex with (E)-2,6-dibromo-4-(2,6-dimethoxystyryl)aniline | | Descriptor: | 2,6-dibromo-4-[(E)-2-(2,6-dimethoxyphenyl)ethenyl]aniline, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
3IN8
 
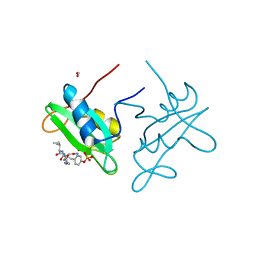 | |
3IMN
 
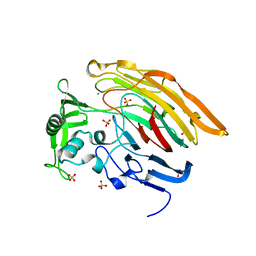 | |
3IMX
 
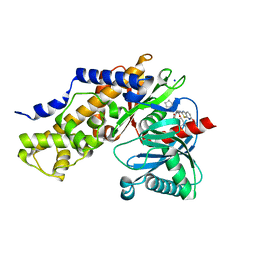 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
3IO7
 
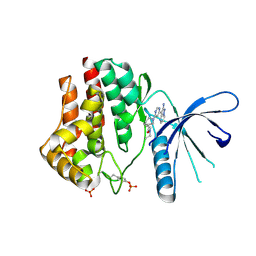 | | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2 | | Descriptor: | (3S)-1-[6-(2-aminopyrazolo[1,5-a]pyrimidin-3-yl)pyrimidin-4-yl]-N,N-diethylpiperidine-3-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IOJ
 
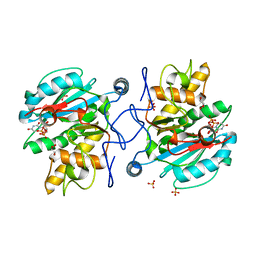 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with UDP | | Descriptor: | GLYCEROL, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Pesnot, T, Jorgensen, R, Palcic, M.M, Wagner, G.K. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and mechanistic basis for a new mode of glycosyltransferase inhibition.
Nat.Chem.Biol., 6, 2010
|
|
3IPW
 
 | |
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IQ5
 
 | |
3IQN
 
 | |
3IQA
 
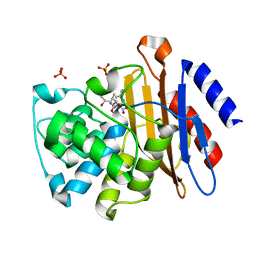 | | Crystal Structure of BlaC covalently bound with Doripenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2009-08-19 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
3I74
 
 | |
3ISP
 
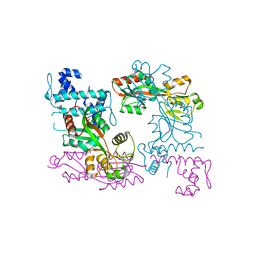 | | Crystal structure of ArgP from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator Rv1985c/MT2039 | | Authors: | Zhou, X, Lou, Z, Sheng, F, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2009-08-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ArgP from Mycobacterium tuberculosis Confirms Two Distinct Conformations of Full-length LysR Transcriptional Regulators and Reveals Its Function in DNA Binding and Transcriptional Regulation.
J.Mol.Biol., 2009
|
|
