7XXL
 
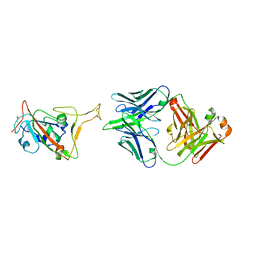 | | RBD in complex with Fab14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | Authors: | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-09-14 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
4ITW
 
 | |
1HNF
 
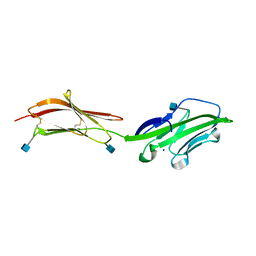 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
3G8A
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoHcy in space group P61 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
5GKA
 
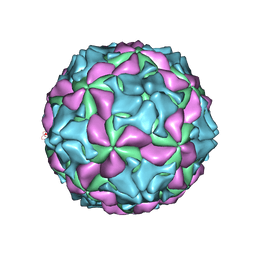 | | cryo-EM structure of human Aichi virus | | Descriptor: | Genome polyprotein, capsid protein VP0, capsid protein VP1 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Tuthill, T.J, Fry, E.E, Rao, Z.H, Stuart, D.I. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of human Aichi virus and implications for receptor binding
Nat Microbiol, 1, 2016
|
|
6BUZ
 
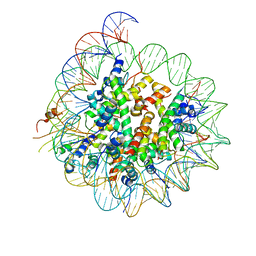 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | Deposit date: | 2017-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
3EKH
 
 | | Calcium-saturated GCaMP2 T116V/K378W mutant monomer | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
1ZHP
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-K505 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z6Z
 
 | | Crystal Structure of Human Sepiapterin Reductase in complex with NADP+ | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Ugochukwu, E, Kavanagh, K, Ng, S, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Sepiapterin Reductase
To be Published
|
|
1TA6
 
 | | Crystal structure of thrombin in complex with compound 14b | | Descriptor: | 1-[2-AMINO-2-CYCLOHEXYL-ACETYL]-PYRROLIDINE-3-CARBOXYLIC ACID 5-CHLORO-2-(2-ETHYLCARBAMOYL-ETHOXY)-BENZYLAMIDE, Hirudin, thrombin | | Authors: | Tucker, T.J, Brady, S.F, Lumma, W.C, Lewis, S.D, Gardel, S.J, Naylor-Olsen, A.M, Yan, Y, Sisko, J.T, Stauffer, K.J, Lucas, B.Y, Lynch, J.J, Cook, J.J, Stranieri, M.T, Holahan, M.A, Lyle, E.A, Baskin, E.P, Chen, I.-W, Dancheck, K.B, Krueger, J.A, Cooper, C.M, Vacca, J.P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-06-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of a series of potent and orally bioavailable
noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position
J.Med.Chem., 41, 1998
|
|
3E3O
 
 | | Glycogen phosphorylase R state-IMP complex | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-08-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycogen phosphorylase revisited: extending the resolution of the R- and T-state structures of the free enzyme and in complex with allosteric activators.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2WO1
 
 | | Crystal Structure of the EphA4 Ligand Binding Domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR, N-PROPANOL | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
1HOO
 
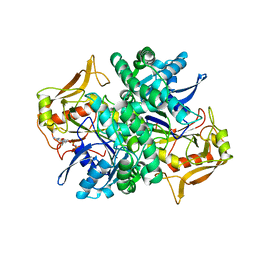 | | STRUCTURE OF GUANINE NUCLEOTIDE (GPPCP) COMPLEX OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI AT PH 6.5 AND 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, AMINOPHOSPHONIC ACID-GUANYLATE ESTER, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Poland, B.W, Hou, Z, Bruns, C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1996-04-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of guanine nucleotide complexes of adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 271, 1996
|
|
3HBN
 
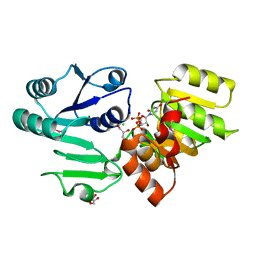 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
5BXV
 
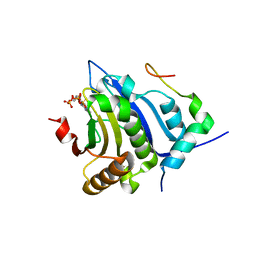 | | eIF4E complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Sekiyama, N, Arthanari, H, Papdopoulos, E, Rodriguez-Mias, R.A, Wagner, G, Leger-Abraham, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6CL2
 
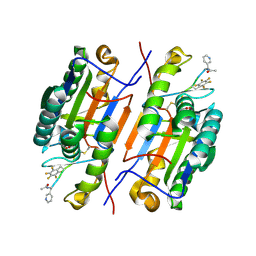 | |
7DFV
 
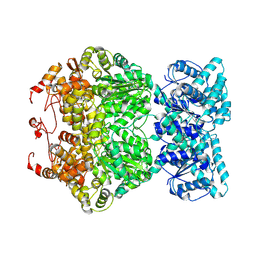 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
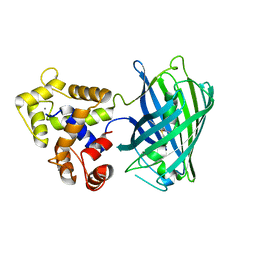
5UKG
 
 | |
4PLU
 
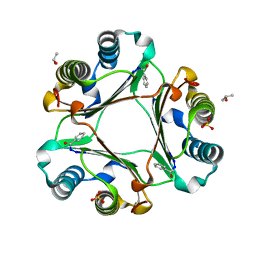 | |
2WUE
 
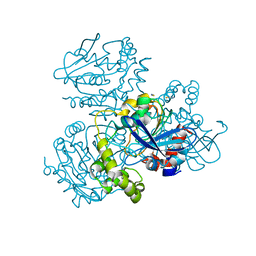 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPODA | | Descriptor: | (3E,5R)-8-(2-CHLOROPHENYL)-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
4NG3
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid
To be Published
|
|
3EHW
 
 | | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Takacs, E, Barabas, O, Vertessy, B.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site
To be Published
|
|
3BIC
 
 | | Crystal structure of human methylmalonyl-CoA mutase | | Descriptor: | CHLORIDE ION, Methylmalonyl-CoA mutase, mitochondrial precursor | | Authors: | Ugochukwu, E, Kochan, G, Pantic, N, Parizotto, E, Pilka, E.S, Pike, A.C.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation.
J.Biol.Chem., 285, 2010
|
|
2CNH
 
 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | CALCIUM ION, N-[(1S)-1-(1H-BENZIMIDAZOL-2-YL)-2-{4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]PHENYL}ETHYL]-4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-SULFONAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
2X07
 
 | | cytochrome c peroxidase: engineered ascorbate binding site | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
