1XGU
 
 | | Structure for antibody HyHEL-63 Y33F mutant complexed with hen egg lysozyme | | Descriptor: | Lysozyme C, antibody kappa heavy chain, antibody kappa light chain | | Authors: | Li, Y, Huang, Y, Swaminathan, C.P, Smith-Gill, S.J, Mariuzza, R.A. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-06 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces
Structure, 13, 2005
|
|
1XDM
 
 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 291K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
1XUF
 
 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE ZINC, CALCIUM ION, TRYPSIN | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XKA
 
 | |
1XKB
 
 | |
1XKR
 
 | | X-ray Structure of Thermotoga maritima CheC | | Descriptor: | chemotaxis protein CheC | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX
Mol.Cell, 16, 2004
|
|
1XMH
 
 | | Structure of Co(II) reconstituted methane monooxygenase hydroxylase from M. capsulatus (Bath) | | Descriptor: | COBALT (II) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Sazinsky, M.H, Merkx, M, Cadieux, E, Tang, S, Lippard, S.J. | | Deposit date: | 2004-10-02 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Preparation and X-ray Structures of Metal-Free, Dicobalt and Dimanganese Forms of Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1XSO
 
 | | THREE-DIMENSIONAL STRUCTURE OF XENOPUS LAEVIS CU,ZN SUPEROXIDE DISMUTASE B DETERMINED BY X-RAY CRYSTALLOGRAPHY AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic Carugo, K, Coda, A, Battistoni, A, Carri, M.T, Polticelli, F, Desideri, A, Rotilio, G, Wilson, K.S, Bolognesi, M. | | Deposit date: | 1995-03-14 | | Release date: | 1995-07-10 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Three-dimensional structure of Xenopus laevis Cu,Zn superoxide dismutase b determined by X-ray crystallography at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1XQN
 
 | | The 15k neutron structure of saccharide-free concanavalin A | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION | | Authors: | Blakeley, M.P, Kalb-Gilboa, A.J, Helliwell, J.R, Myles, D.A.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.5 Å) | | Cite: | The 15-K neutron structure of saccharide-free concanavalin A
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XRM
 
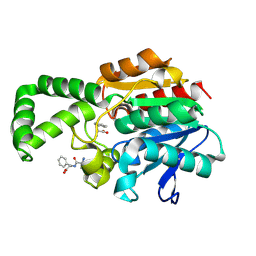 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Ala-Phe | | Descriptor: | ALANINE, PHENYLALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XYF
 
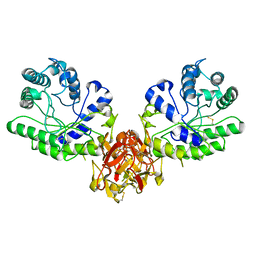 | | ENDO-1,4-BETA-XYLANASE FROM STREPTOMYCES OLIVACEOVIRIDIS | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Fujimoto, Z, Mizuno, H, Kuno, A, Kusakabe, I. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain.
J.Mol.Biol., 300, 2000
|
|
1XXG
 
 | |
1XGI
 
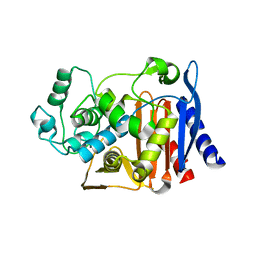 | | AmpC beta-lactamase in complex with 3-(3-nitro-phenylsulfamoyl)-thiophene-2-carboxylic acid | | Descriptor: | 3-{[(3-NITROANILINE]SULFONYL}THIOPHENE-2-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Tondi, D, Morandi, F, Bonnet, R, Costi, M.P, Shoichet, B.K. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based optimization of a non-beta-lactam lead results in inhibitors that do not up-regulate beta-lactamase expression in cell culture.
J.Am.Chem.Soc., 127, 2005
|
|
1XYO
 
 | |
1XHM
 
 | | The Crystal Structure of a Biologically Active Peptide (SIGK) Bound to a G Protein Beta:Gamma Heterodimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1, SIGK Peptide | | Authors: | Davis, T.L, Bonacci, T.M, Smrcka, A.V, Sprang, S.R. | | Deposit date: | 2004-09-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Molecular Characterization of a Preferred Protein Interaction Surface on G Protein betagamma Subunits.
Biochemistry, 44, 2005
|
|
1XZA
 
 | |
1XZJ
 
 | |
1XIW
 
 | | Crystal structure of human CD3-e/d dimer in complex with a UCHT1 single-chain antibody fragment | | Descriptor: | T-cell surface glycoprotein CD3 delta chain, T-cell surface glycoprotein CD3 epsilon chain, immunoglobulin heavy chain variable region, ... | | Authors: | Arnett, K.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YJB
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YFM
 
 | |
2C7P
 
 | | HhaI DNA methyltransferase complex with oligonucleotide containing 2- aminopurine opposite to the target base (GCGC:GMPC) and SAH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*G*GP*AP*TP*GP*(5CM*2PR)*CP*TP*GP*AP*C)-3', 5'-D(*G*TP*CP*AP*GP*CP*GP*CP*AP*TP*CP*C)-3', ... | | Authors: | Neely, R.K, Daujotyte, D, Grazulis, S, Magennis, S.W, Dryden, D.T.F, Klimasauskas, S, Jones, A.C. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-Resolved Fluorescence of 2-Aminopurine as a Probe of Base Flipping in M.HhaI-DNA Complexes.
Nucleic Acids Res., 33, 2005
|
|
1YKL
 
 | | Protocatechuate 3,4-Dioxygenase Y408C mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKK
 
 | | Protocatechuate 3,4-Dioxygenase Y408C Mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YLD
 
 | | Trypsin/BPTI complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-19 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
1YN3
 
 | | Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens | | Descriptor: | truncated cell surface protein map-w | | Authors: | Geisbrecht, B.V, Hamaoka, B.Y, Perman, B, Zemla, A, Leahy, D.J. | | Deposit date: | 2005-01-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens.
J.Biol.Chem., 280, 2005
|
|
