8DR0
 
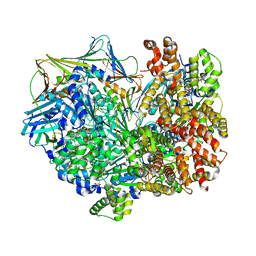 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
6TO1
 
 | |
8E5O
 
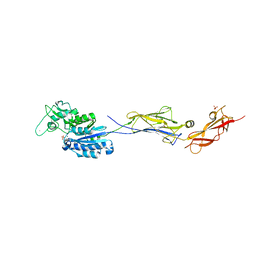
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8DQX
 
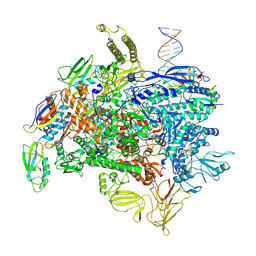
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8E3F
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8DQZ
 
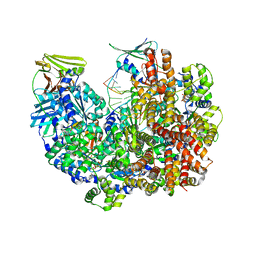
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR6
 
 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
6FOS
 
 | | Cyanidioschyzon merolae photosystem I | | Descriptor: | BETA-CAROTENE, CHLOROPHYLL A, IRON/SULFUR CLUSTER, ... | | Authors: | Nelson, N, Hippler, M, Antoshvili, M, Caspy, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-11 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and function of photosystem I in Cyanidioschyzon merolae.
Photosyn. Res., 139, 2019
|
|
8E5K
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 21 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
6FRI
 
 | |
8EGM
 
 | |
7CVR
 
 | | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine | | Descriptor: | (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2S)-3-phenyl-2-[[(2S)-1-phenylmethoxycarbonylpyrrolidin-2-yl]carbonylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine at 1.60 Angstrom Resolution
To Be Published
|
|
8K58
 
 | | The cryo-EM map of close TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
7CX6
 
 | |
7CX7
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
7CX8
 
 | | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine | | Descriptor: | (1R)-1,2,3,4-tetrahydronaphthalen-1-amine, (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine at 1.70 Angstrom Resolution
To Be Published
|
|
7CKN
 
 | |
7NDJ
 
 | |
7COO
 
 | |
7CON
 
 | |
2H2H
 
 | |
7CP8
 
 | |
7CHL
 
 | | Crystal structure of hybrid Arabinose isomerase AI-10 | | Descriptor: | Hybrid Arabinose isomerase, MANGANESE (II) ION, SODIUM ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of hybrid Arabinose isomerase AI-10
To Be Published
|
|
6UM2
 
 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
