8U16
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
3P91
 
 | |
2H79
 
 | | Crystal Structure of human TR alpha bound T3 in orthorhombic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
8UN9
 
 | |
3DZD
 
 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
1NEX
 
 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
8TZX
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | Descriptor: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
8UV1
 
 | | Structure of TDP1 complexed with compound IB01 | | Descriptor: | 1,2-ETHANEDIOL, 8-(fluorosulfonyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke, T.R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of TDP1 complexed with compound IB01
To Be Published
|
|
7E1L
 
 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
6FZJ
 
 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
2UV1
 
 | |
7DA2
 
 | | The crystal structure of the chicken FANCM-MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Nishino, T, Ito, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analysis of the chicken FANCM-MHF complex and its stability.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6FZY
 
 | | PPAR mutant | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Rochel, N. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
6FZG
 
 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, Peroxisome proliferator-activated receptor gamma | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZF
 
 | | PPAR mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZP
 
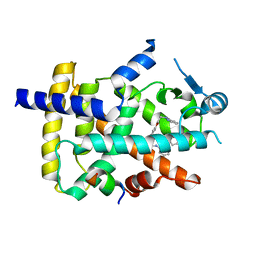 | | PPAR gamma complex. | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Rochel, N, Beji, S. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
4ML0
 
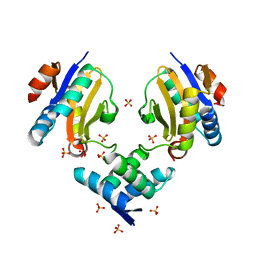 | | Crystal structure of E.coli DinJ-YafQ complex | | Descriptor: | Predicted antitoxin of YafQ-DinJ toxin-antitoxin system, Predicted toxin of the YafQ-DinJ toxin-antitoxin system, SULFATE ION | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
2RT6
 
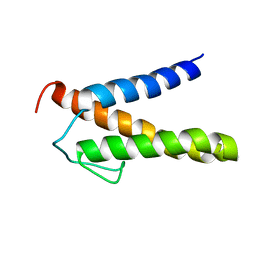 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for PriC N-terminal domain | | Descriptor: | Primosomal replication protein N'' | | Authors: | Aramaki, T, Abe, Y, Katayama, T, Ueda, T. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a replication restart primosome factor, PriC, in Escherichia coli.
Protein Sci., 22, 2013
|
|
3IH2
 
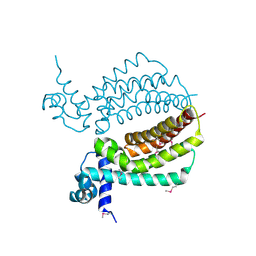 | | TM1030 crystallized at 323K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
3IH4
 
 | | TM1030 crystallized at 277K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
3IH3
 
 | | TM1030 crystallized at 310K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
8WH3
 
 | | MPOX E5 hexamer apo form | | Descriptor: | Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
6O7H
 
 | |
