2YBM
 
 | |
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
2YPZ
 
 | |
2YBJ
 
 | |
2YKP
 
 | |
2YJE
 
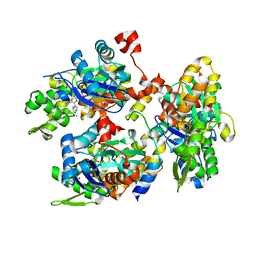 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
2YKQ
 
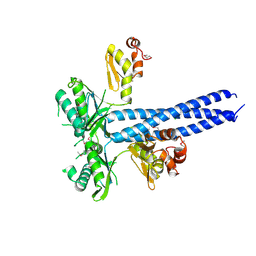 | |
2YMT
 
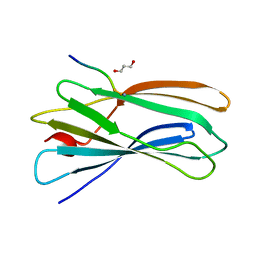 | | gamma 2 adaptin EAR domain crystal structure with phage peptide GEEWGPWV | | Descriptor: | 1,3-PROPANDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, PHAGE DISPLAY DERIVED GAMMA 2 ADAPTIN EAR DOMAIN BINDING PEPTIDE | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-10-10 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
2YQ0
 
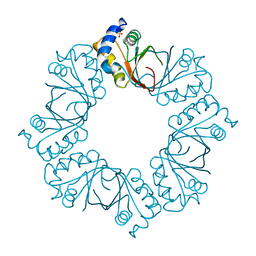 | |
2Y78
 
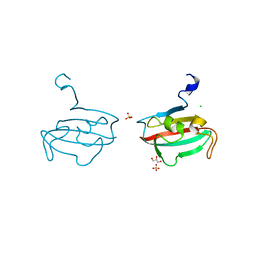 | | Crystal structure of BPSS1823, a Mip-like chaperone from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Norville, I.H, O'Shea, K, Sarkar-Tyson, M, Harmer, N.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | The Structure of a Burkholderia Pseudomallei Immunophilin-Inhibitor Complex Reveals New Approaches to Antimicrobial Development
Biochem.J., 437, 2011
|
|
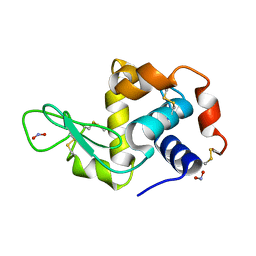
2YBL
 
 | |
2YKO
 
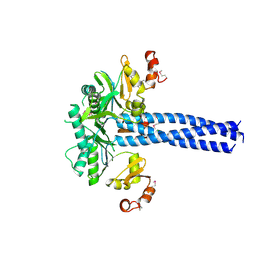 | |
2YC1
 
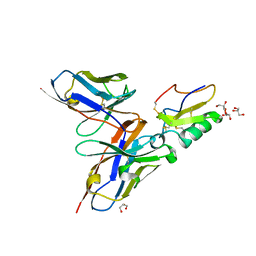 | | Crystal structure of the human derived single chain antibody fragment (scFv) 9004G in complex with Cn2 toxin from the scorpion Centruroides noxius Hoffmann | | Descriptor: | BETA-MAMMAL TOXIN CN2, GLYCEROL, SINGLE CHAIN ANTIBODY FRAGMENT 9004G | | Authors: | Canul-Tec, J.C, Riano-Umbarila, L, Rudino-Pinera, E, Becerril, B, Possani, L.D, Torres-Larios, A. | | Deposit date: | 2011-03-10 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Neutralization of the Major Toxic Component from the Scorpion Centruroides Noxius Hoffmann by a Human-Derived Single Chain Antibody Fragment.
J.Biol.Chem., 286, 2011
|
|
2Y98
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in P21212 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-02-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2YBH
 
 | |
2YBR
 
 | | Crystal structure of the human derived single chain antibody fragment (scFv) 9004G in complex with Cn2 toxin from the scorpion Centruroides noxius Hoffmann | | Descriptor: | BETA-MAMMAL TOXIN CN2, SINGLE CHAIN ANTIBODY FRAGMENT 9004G | | Authors: | Canul-Tec, J.C, Riano-Umbarila, L, Rudino-Pinera, E, Becerril, B, Possani, L.D, Torres-Larios, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Neutralization of the Major Toxic Component from the Scorpion Centruroides Noxius Hoffmann by a Human-Derived Single Chain Antibody Fragment
J.Biol.Chem., 286, 2011
|
|
3BBZ
 
 | | Structure of the nucleocapsid-binding domain from the mumps virus phosphoprotein | | Descriptor: | BROMIDE ION, FORMIC ACID, P protein | | Authors: | Kingston, R.L, Gay, L.S, Baase, W.S, Matthews, B.W. | | Deposit date: | 2007-11-11 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization
J.Mol.Biol., 379, 2008
|
|
3BJ5
 
 | | Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b' domain | | Descriptor: | Protein disulfide-isomerase, SULFATE ION | | Authors: | Ruddock, L.W, Nguyen, V.D, Wierenga, R.K, Haapalainen, A.M. | | Deposit date: | 2007-12-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b' domain
J.Mol.Biol., 383, 2008
|
|
3B8I
 
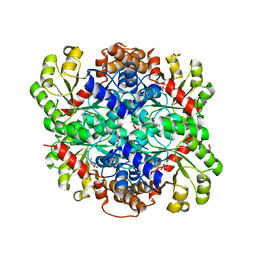 | |
3B4O
 
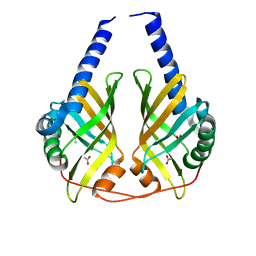 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3B0F
 
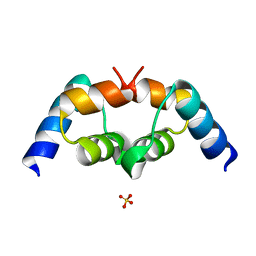 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZUQ
 
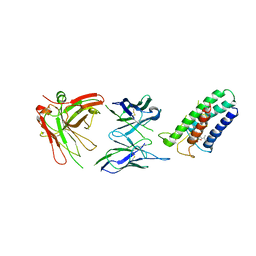 | | Crystal structure of DsbB-Fab complex | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
3B47
 
 | | Periplasmic sensor domain of chemotaxis protein GSU0582 | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
3B4P
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | Descriptor: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
