5OKH
 
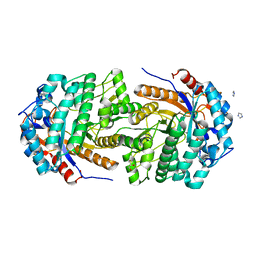 | | Conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in the C2 spacegroup | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
8RZJ
 
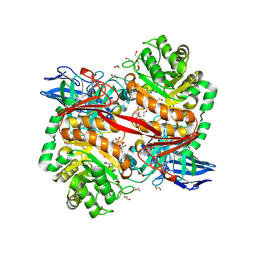 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | Descriptor: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5J42
 
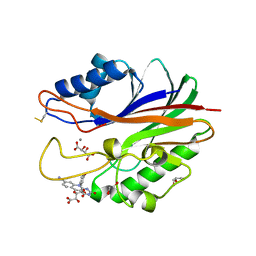 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 10-(4-hydroxyphenyl)-2,4-dioxo-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, GLYCEROL, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
6FYI
 
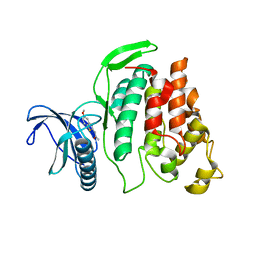 | | X-ray Structure of CLK2-KD(130-496)/TG003 at 2.6A | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
5MG9
 
 | | Putative Ancestral Mamba toxin 1 (AncTx1-W28R/I38S) | | Descriptor: | 1,2-ETHANEDIOL, AncTx1-W28R/I38S, S-1,2-PROPANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Blanchet, G, Mourier, G, Servent, D. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins.
Sci Rep, 7, 2017
|
|
5TGC
 
 | | Structure of the hetero-trimer of Rtt102-Arp7/9 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Turegun, B, Dominguez, R. | | Deposit date: | 2016-09-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Actin-related proteins regulate the RSC chromatin remodeler by weakening intramolecular interactions of the Sth1 ATPase.
Commun Biol, 1, 2018
|
|
9JRM
 
 | | outward-open hSLC19A1 + 2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Soluble cytochrome b562,Reduced folate transporter,Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
5OKJ
 
 | | Non-conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in the C2 spacegroup | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
7NRZ
 
 | | Crystal structure of malate dehydrogenase from Trypanosoma cruzi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sonani, R.R, Kurpiewska, K, Lewinski, K, Dubin, G. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct sequence and structural feature of trypanosoma malate dehydrogenase.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
5U25
 
 | |
5ZJV
 
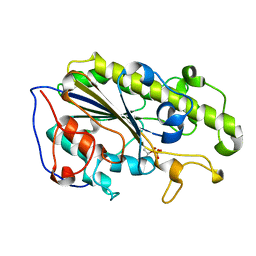 | | Crystal structure of the catalytic domain of MCR-1 (cMCR-1) in complex with xylose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-L-xylopyranose | | Authors: | Liu, Z.X, Han, Z, Yu, X.L, Wen, G, Zeng, C. | | Deposit date: | 2018-03-22 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the Catalytic Domain of MCR-1 (cMCR-1) in Complex with d-Xylose
Crystals, 8, 2018
|
|
8JNR
 
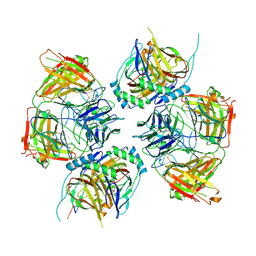 | |
9JZ2
 
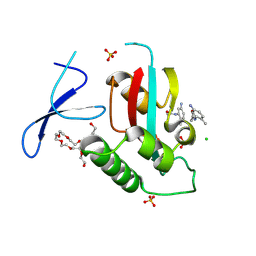 | | Crystal structure of the PIN1 and fragment 7 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, N-(5-azanyl-2-methyl-phenyl)ethanamide, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-10-13 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9JYP
 
 | | Crystal structure of the PIN1 and fragment 3 complex | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-oxidanylidene-4-thiophen-2-yl-butanoic acid, CHLORIDE ION, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-10-12 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KEQ
 
 | | Crystal structure of the PIN1 and fragment 27 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3-(4-methylpiperazin-1-yl)aniline, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
5IA5
 
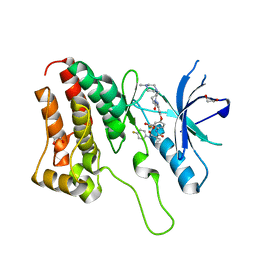 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with golvatinib (E7050) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, golvatinib | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
9KFC
 
 | | Crystal structure of the PIN1 and fragment 40 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9JYO
 
 | | Crystal structure of the Pin1 and fragment 2 complex. | | Descriptor: | 2-(4-aminophenyl)ethanoic acid, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-10-12 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
5O5F
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ038 | | Descriptor: | 1-ethyl-~{N}-[(~{R})-(3-fluorophenyl)-(1-methylimidazol-2-yl)methyl]-2,3-bis(oxidanylidene)-4~{H}-quinoxaline-6-carboxamide, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
9JZ3
 
 | | Crystal structure of the PIN1 and fragment 8 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3H-benzimidazol-5-ylmethanol, CHLORIDE ION, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-10-13 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KF0
 
 | | Crystal structure of the PIN1 and fragment 32 complex. | | Descriptor: | (2,5-dimethylpyrazol-3-yl)methanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KXQ
 
 | | Crystal structure of the PIN1 and fragment 55 complex. | | Descriptor: | (3,5-dimethyl-1,2-oxazol-4-yl)methanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-12-07 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
6BYF
 
 | |
9NST
 
 | | Bacterial Pictet-Spenglerase KslB in complex with product of L-Trp and a-ketoglutaric acid | | Descriptor: | (1~{S},3~{S})-1-(3-hydroxy-3-oxopropyl)-2,3,4,9-tetrahydropyrido[3,4-b]indole-1,3-dicarboxylic acid, Pictet-Spenglerase | | Authors: | Kim, K, Kim, W. | | Deposit date: | 2025-03-17 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and mechanistic insights into KslB, a bacterial Pictet-Spenglerase in kitasetaline biosynthesis.
Rsc Chem Biol, 6, 2025
|
|
6UFZ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
