1RKA
 
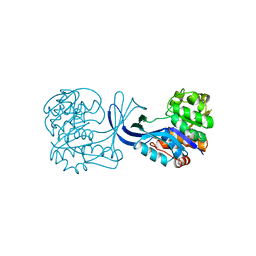 | |
1JIT
 
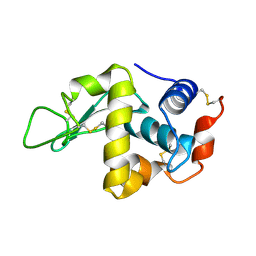 | |
1K12
 
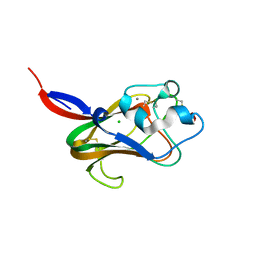 | | Fucose Binding lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, LECTIN, ... | | Authors: | Bianchet, M.A, Odom, E.W, Vasta, G.R, Amzel, L.M. | | Deposit date: | 2001-09-23 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel fucose recognition fold involved in innate immunity.
Nat.Struct.Biol., 9, 2002
|
|
1JKD
 
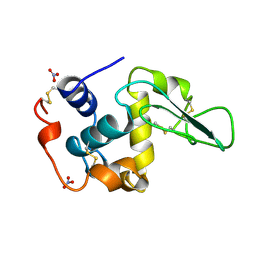 | | HUMAN LYSOZYME MUTANT WITH TRP 109 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
3RSK
 
 | |
1L1W
 
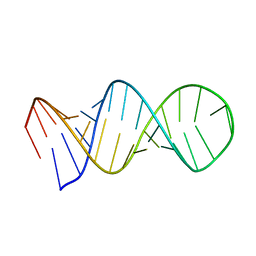 | | NMR structure of a SRP19 binding domain in human SRP RNA | | Descriptor: | SRP19 binding domain of SRP RNA | | Authors: | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
2C11
 
 | | Crystal structure of the 2-hydrazinopyridine of semicarbazide- sensitive amine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jakobsson, E, Kleywegt, G.J. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human semicarbazide-sensitive amine oxidase/vascular adhesion protein-1.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
6K4V
 
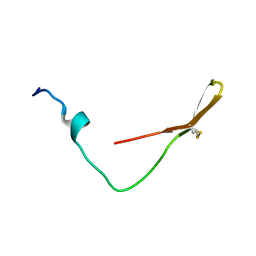 | | The solution structure of the smart chimeric peptide G6 | | Descriptor: | smart chimeric peptide G6 | | Authors: | Wang, J.H, Liu, X.H. | | Deposit date: | 2019-05-27 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
1K4K
 
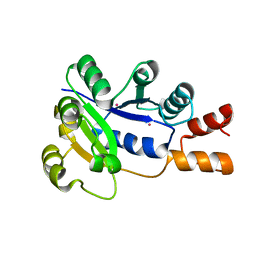 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
1UQR
 
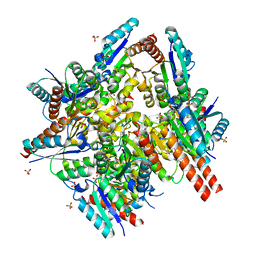 | | Type II 3-dehydroquinate dehydratase (DHQase) from Actinobacillus pleuropneumoniae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Maes, D, Gonzalez-Ramirez, L.A, Lopez-Jaramillo, J, Yu, B, De Bondt, H, Zegers, I, Afonina, E, Garcia-Ruiz, J.M, Gulnik, S. | | Deposit date: | 2003-10-16 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Study of the Type II 3-Dehydroquinate Dehydratase from Actinobacillus Pleuropneumoniae
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7CM3
 
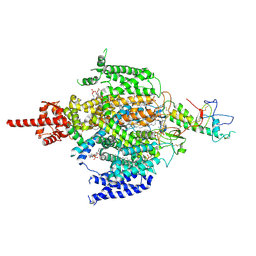 | | Cryo-EM structure of human NALCN in complex with FAM155A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J, Yan, Z, Ke, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human sodium leak channel NALCN in complex with FAM155A.
Nat Commun, 11, 2020
|
|
1KBG
 
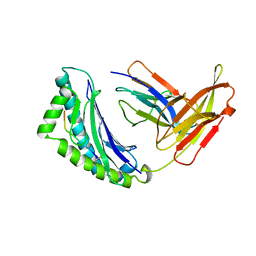 | | MHC Class I H-2KB Presented Glycopeptide RGY8-6H-GAL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN), ... | | Authors: | Speir, J.A, Abdel-Motal, U.M, Jondal, M, Wilson, I.A. | | Deposit date: | 1998-08-28 | | Release date: | 1999-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL.
Immunity, 10, 1999
|
|
6HRQ
 
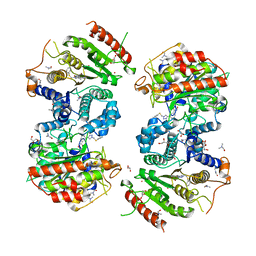 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-09-28 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTT
 
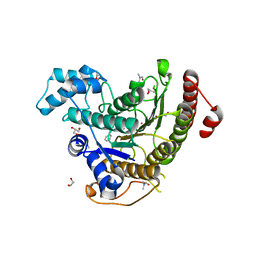 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 7 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3EOG
 
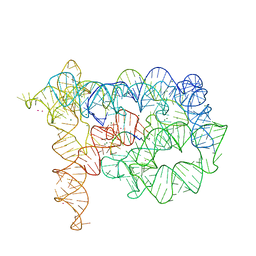 | | Co-crystallization showing exon recognition by a group II intron | | Descriptor: | 5'-R(*UP*UP*AP*UP*UP*A)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Rajashankar, K, Keating, K.S, Pyle, A.M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Structural basis for exon recognition by a group II intron.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6HTH
 
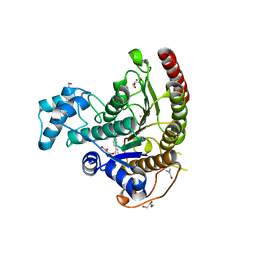 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 5 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-[(4-phenylphenyl)carbonylamino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU3
 
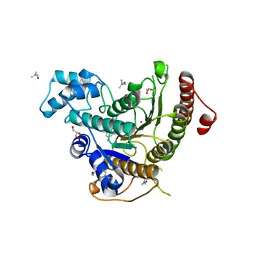 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor | | Descriptor: | 1-[5-chloranyl-2-(4-fluoranylphenoxy)phenyl]-~{N}-oxidanyl-1,2,3-triazole-4-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
1UR9
 
 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
6ZX6
 
 | |
6ZX7
 
 | |
1KQQ
 
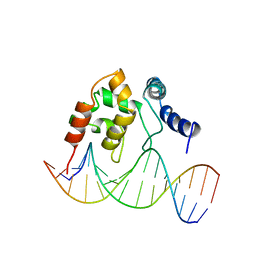 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1LAT
 
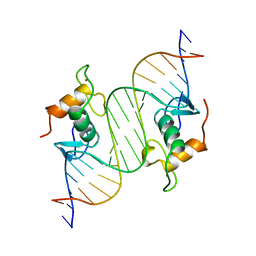 | | GLUCOCORTICOID RECEPTOR MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*TP*TP*CP*TP*G P*GP*A)-3'), GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Gewirth, D.T, Sigler, P.B. | | Deposit date: | 1995-12-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex.
Nat.Struct.Biol., 2, 1995
|
|
1JJ3
 
 | |
1JKO
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*CP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*GP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
6G3B
 
 | | AvaII restriction endonuclease in complex with an RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*GP*TP*CP*CP*TP*A)-3'), RNA (5'-R(P*GP*UP*AP*GP*GP*AP*CP*CP*AP*UP*G)-3'), Type II site-specific deoxyribonuclease | | Authors: | Kisiala, M, Kowalska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2018-03-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation
Nucleic Acids Res., 2020
|
|
