7H09
 
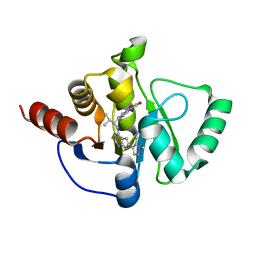 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011177-001 | | Descriptor: | (4M)-1-methyl-4-(4-{[(1R)-2-methyl-1-(3-oxo-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0Q
 
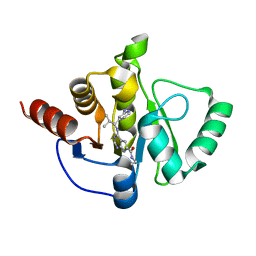 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013821-001 | | Descriptor: | 4-{[(1S)-1-(2-aminopyridin-4-yl)-2-methylpropyl]amino}-N-ethyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4ZJM
 
 | | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein LpqH, ... | | Authors: | Arbing, M.A, Chan, S, Kuo, E, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763)
To Be Published
|
|
4WHE
 
 | | Crystal structure of E. coli phage shock protein A (PspA 1-144) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phage shock protein A | | Authors: | Parthier, C, Schoepfel, M, Stubbs, M.T, Osadnik, H, Brueser, T. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Mol.Microbiol., 98, 2015
|
|
4OQG
 
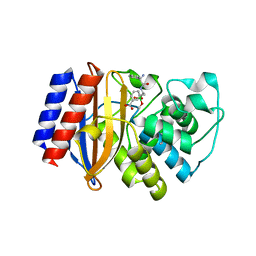 | | Crystal structure of TEM-1 beta-lactamase in complex with boron-based inhibitor EC25 | | Descriptor: | 3-[(2R)-2-{[(2R)-2-amino-2-phenylacetyl]amino}-2-(dihydroxyboranyl)ethyl]benzoic acid, Ampicillin resistance protein, ZINC ION | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-09 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7GZ1
 
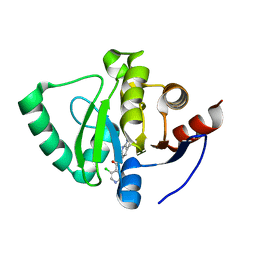 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000061-001 | | Descriptor: | 3-chloro-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)pyridine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ0
 
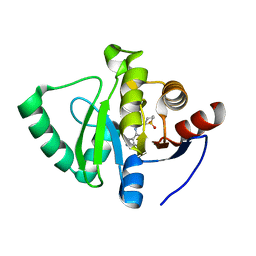 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000051-001 | | Descriptor: | (3S)-3-{[(1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]methyl}-1lambda~6~-thiane-1,1-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3ENS
 
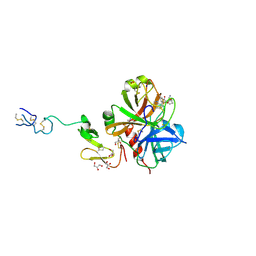 | |
7GZE
 
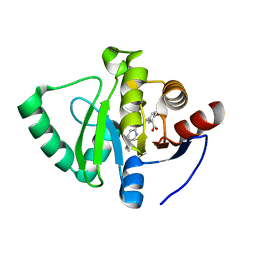 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000601-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-{[5-(dimethylamino)pyridine-2-carbonyl]amino}propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZM
 
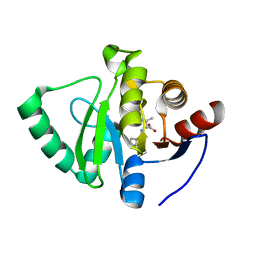 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008324-001 | | Descriptor: | N-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-D-valine, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4WK5
 
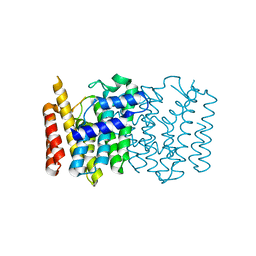 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
1ANA
 
 | |
1O91
 
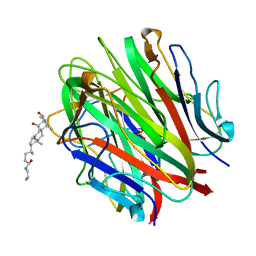 | | Crystal Structure of a Collagen VIII NC1 Domain Trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, COLLAGEN ALPHA 1(VIII) CHAIN, SULFATE ION | | Authors: | Kvansakul, M, Bogin, O, Hohenester, E, Yayon, A. | | Deposit date: | 2002-12-10 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Collagen Alpha1(Viii) Nc1 Trimer.
Matrix Biol., 22, 2003
|
|
7H0F
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011207-001 | | Descriptor: | N-cyclopropyl-4-{[(1R)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidine-5-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
8SCX
 
 | |
7H0B
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0010716-001 | | Descriptor: | (4M)-4-(4-{[(1R)-1-(2,3-dihydro[1,4]dioxino[2,3-b]pyridin-6-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0Y
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013258-001 | | Descriptor: | 4-{[(1S)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-N-(1-methylazetidin-3-yl)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
2BKT
 
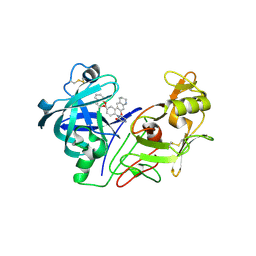 | | crystal structure of renin-pf00257567 complex | | Descriptor: | 1-{4-[3-(2-METHOXY-BENZYLOXY)-PROPOXY]-PHENYL}-6-(1,2,,3,4-TETRAHYDRO-QUINOLIN-7-YLOXYMETHYL)-PIPERAZIN-2-ONE, RENIN | | Authors: | Powell, N.A, Clay, E.H, Holsworth, D.D, Edmunds, J.J, Bryant, J.W, Ryan, J.M, Jalaie, M, Zhang, E. | | Deposit date: | 2005-02-18 | | Release date: | 2006-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Equipotent Activity in Both Enantiomers of a Series of Ketopiperazine-Based Renin Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6RKQ
 
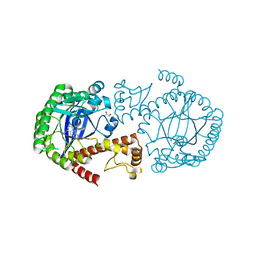 | | Crystal Structure of TGT in complex with N2-methyl-8-(prop-1-yn-1-yl)-3H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | (8~{R})-~{N}2-methyl-8-prop-1-ynyl-7,8-dihydro-3~{H}-imidazo[4,5-g]quinazoline-2,6-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
7H0S
 
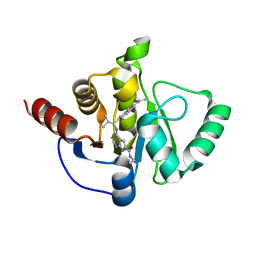 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013769-001 | | Descriptor: | 4-{[(1S)-1-(2-acetamido-1,3-benzothiazol-6-yl)-2-methylpropyl]amino}-N-ethyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | Descriptor: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-12 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
7DJI
 
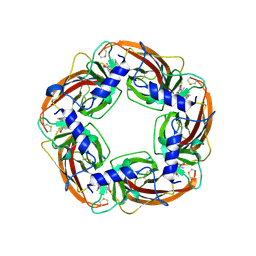 | |
2R4Z
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: Structure of I25W with CO | | Descriptor: | CARBON MONOXIDE, Globin-1, PHOSPHATE ION, ... | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
7GZC
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000588-001 | | Descriptor: | (3R)-3-(4-fluorophenyl)-3-[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.185 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
