3QYT
 
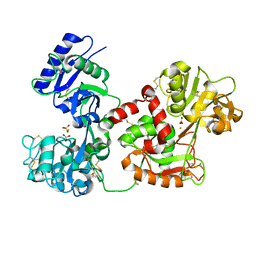 | | Diferric bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2011-03-03 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe
Sci Rep, 2, 2012
|
|
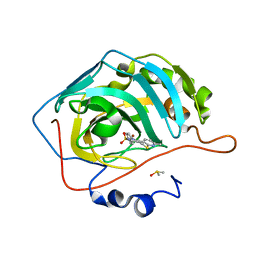
3QYK
 
 | |
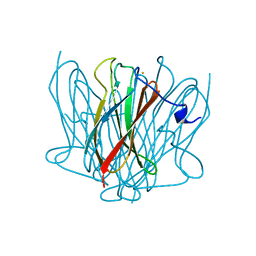
3QZB
 
 | |
3QZS
 
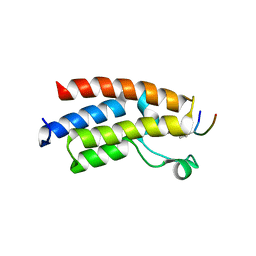 | |
3PVR
 
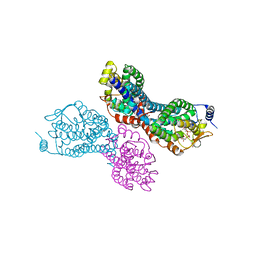 | |
3PWF
 
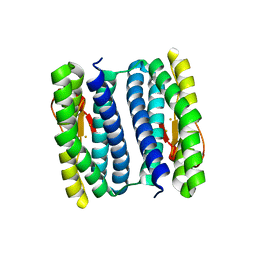 | | High resolution structure of the fully reduced form of rubrerythrin from P. furiosus | | Descriptor: | FE (II) ION, Rubrerythrin | | Authors: | Dillard, B.D, Demick, J.M, Adams, M.W, Lanzilotta, W.N. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
3PWP
 
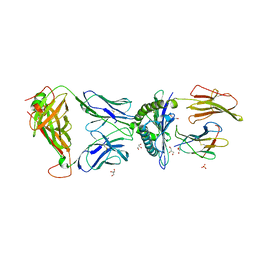 | |
3PWX
 
 | | Structure of putative flagellar hook-associated protein from Vibrio parahaemolyticus | | Descriptor: | MAGNESIUM ION, Putative flagellar hook-associated protein | | Authors: | Ramagopal, U.A, Patskovsky, Y, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-09 | | Release date: | 2011-01-19 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of putative flagellar hook-associated protein from Vibrio parahaemolyticus
To be published
|
|
3Q0G
 
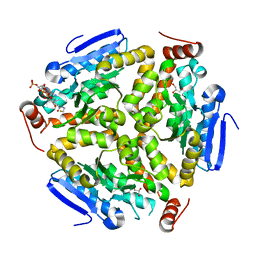 | | Crystal Structure of the Mycobacterium tuberculosis Crotonase Bound to a Reaction Intermediate Derived from Crotonyl CoA | | Descriptor: | Butyryl Coenzyme A, COENZYME A, GLYCEROL, ... | | Authors: | Bruning, J.B, Delgado, E, Ghosh, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the Prokaryotic Crotonase
To be Published
|
|
3Q1N
 
 | |
3PZD
 
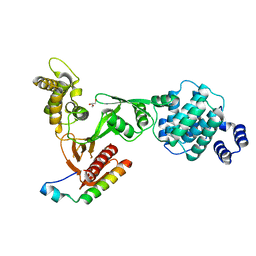 | | Structure of the myosin X MyTH4-FERM/DCC complex | | Descriptor: | GLYCEROL, Myosin-X, Netrin receptor DCC | | Authors: | Wei, Z, Yan, J, Pan, L, Zhang, M. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q12
 
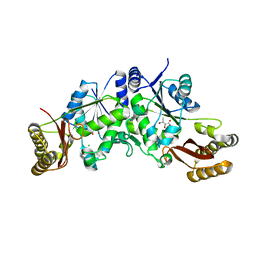 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
3PKA
 
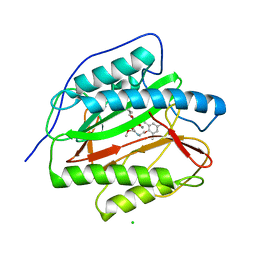 | | M. tuberculosis MetAP with bengamide analog Y02, in Mn form | | Descriptor: | (2R,3R,4S,5R,6E)-3,4,5-trihydroxy-2-methoxy-8,8-dimethyl-N-[2-(2,4,6-trimethylphenoxy)ethyl]non-6-enamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
3PL0
 
 | |
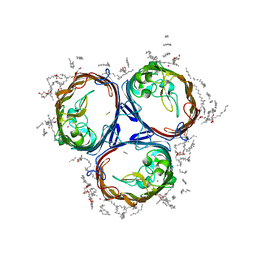
3POX
 
 | |
3PPY
 
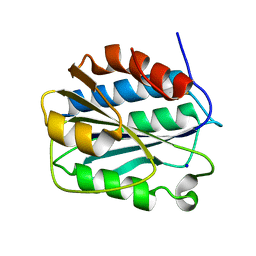 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3Q2X
 
 | |
3PRP
 
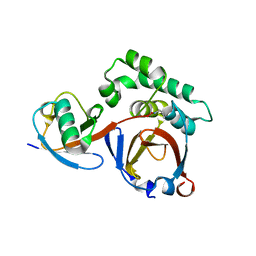 | | Structural analysis of a viral OTU domain protease from the Crimean-Congo Hemorrhagic Fever virus in complex with human ubiquitin | | Descriptor: | Polyubiquitin-B (Fragment), RNA-directed RNA polymerase L | | Authors: | Capodagli, G.C, McKercher, M.A, Baker, E.A, Masters, E.M, Brunzelle, J.S, Pegan, S.D. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin.
J.Virol., 85, 2011
|
|
3Q3M
 
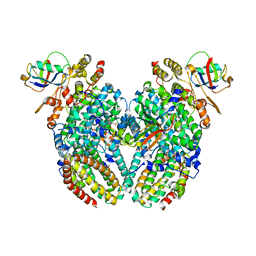 | | Toluene 4 monooxygenase HD Complex with Inhibitor 4-Bromobenzoate | | Descriptor: | 4-bromobenzoic acid, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3PTO
 
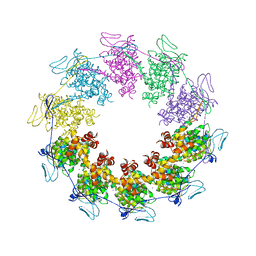 | |
3PUH
 
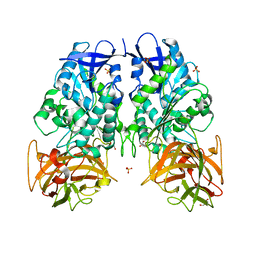 | |
3PW3
 
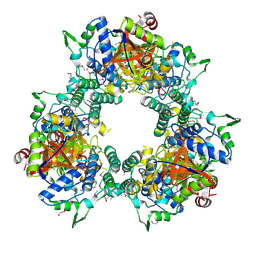 | |
3Q0Y
 
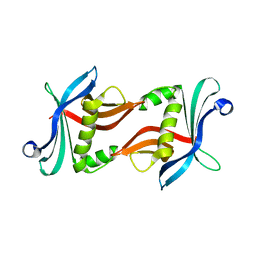 | | N-terminal domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
3Q3Z
 
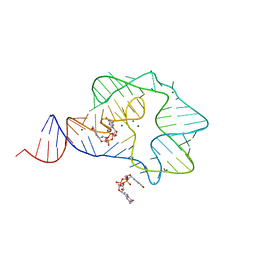 | | Structure of a c-di-GMP-II riboswitch from C. acetobutylicum bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, c-di-GMP-II riboswitch | | Authors: | Smith, K.D, Shanahan, C.A, Moore, E.L, Simon, A.C, Strobel, S.A. | | Deposit date: | 2010-12-22 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of differential ligand recognition by two classes of bis-(3'-5')-cyclic dimeric guanosine monophosphate-binding riboswitches.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q6W
 
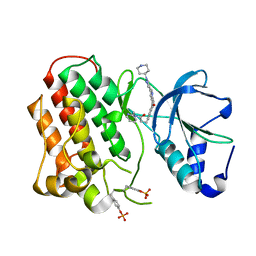 | | Structure of dually-phosphorylated MET receptor kinase in complex with an MK-2461 analog with specificity for the activated receptor | | Descriptor: | 3-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)propanamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
