7LPH
 
 | | APE1 Mn-bound product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7N8K
 
 | | LINE-1 endonuclease domain complex with Mg | | Descriptor: | ACETATE ION, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7N94
 
 | | LINE-1 endonuclease domain complex with DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*CP*TP*TP*AP*AP*AP*AP*AP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*TP*CP*CP*TP*TP*TP*TP*TP*AP*AP*GP*GP*GP*CP*TP*A)-3'), LINE-1 retrotransposable element ORF2 protein | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7N3Z
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
7N3Y
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
3G2C
 
 | | Mth0212 in complex with a short ssDNA (CGTA) | | Descriptor: | 5'-D(P*CP*GP*TP*A)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G4T
 
 | | Mth0212 (WT) in complex with a 7bp dsDNA | | Descriptor: | 5'-D(*CP*G*TP*AP*CP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-04 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G3C
 
 | | Mth0212 (WT) in complex with a 6bp dsDNA containing a single one nucleotide long 3'-overhang | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G3Y
 
 | | Mth0212 in complex with ssDNA in space group P32 | | Descriptor: | 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-03 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G2D
 
 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3GA6
 
 | | Mth0212 in complex with two DNA helices | | Descriptor: | 5'-D(*GP*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G91
 
 | | 1.2 Angstrom structure of the exonuclease III homologue Mth0212 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-12 | | Release date: | 2010-03-09 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA.
J.Mol.Biol., 399, 2010
|
|
3FZI
 
 | |
3G38
 
 | | The catalytically inactive mutant Mth0212 (D151N) in complex with an 8 bp dsDNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*GP*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G0R
 
 | | Complex of Mth0212 and an 8bp dsDNA with distorted ends | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G1K
 
 | |
3G00
 
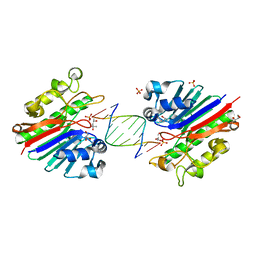 | | Mth0212 in complex with a 9bp blunt end dsDNA at 1.7 Angstrom | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*GP*TP*AP*TP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*UP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G0A
 
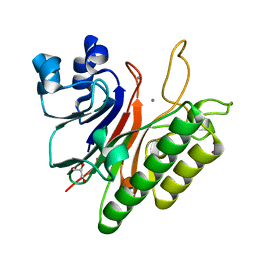 | | Mth0212 with two bound manganese ions | | Descriptor: | Exodeoxyribonuclease, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G6S
 
 | | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus. Northeast Structural Genomics Consortium Target BvR56D | | Descriptor: | Putative endonuclease/exonuclease/phosphatase family protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Xiao, R, Sahdev, S, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus.
To be Published
|
|
3G8V
 
 | |
7AX1
 
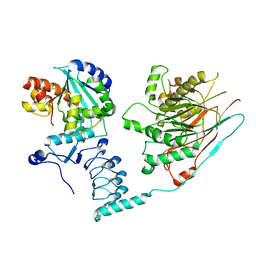 | | Crystal structure of the human CCR4-CAF1 complex | | Descriptor: | CCR4-NOT transcription complex subunit 6, CCR4-NOT transcription complex subunit 7, MAGNESIUM ION | | Authors: | Chen, Y, Khazina, E, Weichenrieder, O. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex.
Nucleic Acids Res., 49, 2021
|
|
1SR4
 
 | | Crystal Structure of the Haemophilus ducreyi cytolethal distending toxin | | Descriptor: | BROMIDE ION, Cytolethal distending toxin subunit A, cytolethal distending toxin protein B, ... | | Authors: | Nesic, D, Hsu, Y, Stebbins, C.E. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assembly and Function of a Bacterial Genotoxin
Nature, 429, 2004
|
|
7VOI
 
 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
6RW0
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase | | Descriptor: | GLYCEROL, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
6RVZ
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase, in complex with adenosine-2',3'-vanadate | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
