7R50
 
 | |
5EBG
 
 | | Crystal structure of bovine CD8aa homodimer | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain | | Authors: | Liu, Y, Li, X, Zhang, N, Qi, J, Xia, C. | | Deposit date: | 2015-10-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
8EQ8
 
 | |
8EQH
 
 | |
5DRV
 
 | |
6HFJ
 
 | |
6HFS
 
 | |
4JLT
 
 | | Crystal structure of P450 2B4(H226Y) in complex with paroxetine | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, GLYCEROL, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Structural Snapshot of CYP2B4 in Complex with Paroxetine Provides Insights into Ligand Binding and Clusters of Conformational States.
J.Pharmacol.Exp.Ther., 346, 2013
|
|
5DNI
 
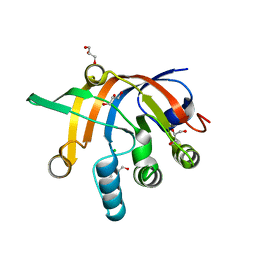 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayaraman, V, Kunala, J, Balaram, H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
7RA7
 
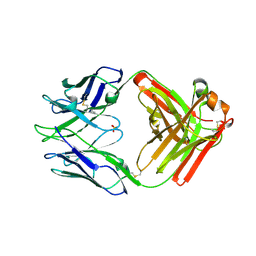 | | Crystal structure of rabbit anti-HIV Fab 11A | | Descriptor: | 11A Fab heavy chain, 11A Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Limited breadth of anti-HIV Env glycan hole antibodies is further hindered by strain-specific peptide interactions
To Be Published
|
|
5E6N
 
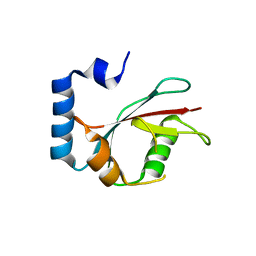 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
4KLB
 
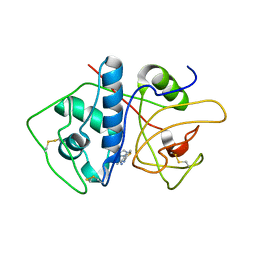 | | Crystal Structure of Cruzain in complex with the non-covalent inhibitor Nequimed176 | | Descriptor: | 2-{[(1H-1,2,4-triazol-5-ylsulfanyl)acetyl]amino}thiophene-3-carboxamide, Cruzipain | | Authors: | Fernandes, W.B, Montanari, C.A, Mckerrow, J.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Non-peptidic Cruzain Inhibitors with Trypanocidal Activity Discovered by Virtual Screening and In Vitro Assay.
Plos Negl Trop Dis, 7, 2013
|
|
2RAA
 
 | |
4KQ8
 
 | | Structure of Recombinant Human Cytochrome P450 Aromatase | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Cytochrome P450 19A1, PHOSPHATE ION, ... | | Authors: | Ghosh, D, Di Nardo, G, Griswold, J. | | Deposit date: | 2013-05-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the functional roles of critical residues in human cytochrome p450 aromatase.
Biochemistry, 52, 2013
|
|
8F70
 
 | |
8F5N
 
 | | Identification of an Immunodominant region on a Group A Streptococcus T-antigen Reveals Temperature-Dependent Motion in Pili | | Descriptor: | CALCIUM ION, Mouse-Human Fab heavy chain, Mouse-Human Fab light chain, ... | | Authors: | Raynes, J.M, Young, P.G, Moreland, N.J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of an immunodominant region on a group A Streptococcus T-antigen reveals temperature-dependent motion in pili.
Virulence, 14, 2023
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
5E24
 
 | | Structure of the Su(H)-Hairless-DNA Repressor Complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Kovall, R.A, Yuan, Z. | | Deposit date: | 2015-09-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and Function of the Su(H)-Hairless Repressor Complex, the Major Antagonist of Notch Signaling in Drosophila melanogaster.
Plos Biol., 14, 2016
|
|
4P86
 
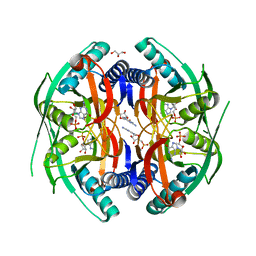 | | Structure of PyrR protein from Bacillus subtilis with GMP | | Descriptor: | Bifunctional protein PyrR, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
7R58
 
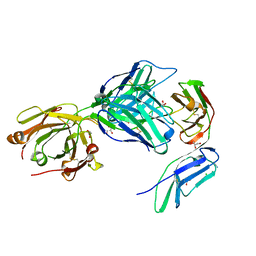 | | Crystal structure of the GPVI-glenzocimab complex | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Jandrot-Perrus, M, Lebozec, K, Rose, N, Welin, M, Billiald, P. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition.
Blood Adv, 7, 2023
|
|
4K22
 
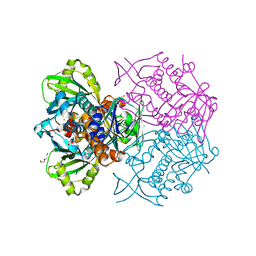 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
8ER1
 
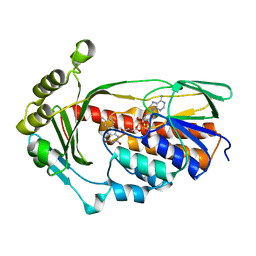 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ER0
 
 | | X-ray crystal structure of Tet(X6) bound to anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ESD
 
 | | Crystal structure of COMMD7-COMMD9-COMMD5-COMMD10 tetramer | | Descriptor: | COMM domain-containing protein 10, COMM domain-containing protein 5, COMM domain-containing protein 7, ... | | Authors: | Healy, M.D, Collins, B.M. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8ESE
 
 | |
