4G51
 
 | | Crystallographic analysis of the interaction of nitric oxide with hemoglobin from Trematomus bernacchii in the T quaternary structure (fully ligated state). | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRIC OXIDE, ... | | Authors: | Merlino, A, Balsamo, A, Pica, A, Mazzarella, L, Vergara, A. | | Deposit date: | 2012-07-17 | | Release date: | 2013-01-16 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective X-ray-induced NO photodissociation in haemoglobin crystals: evidence from a Raman-assisted crystallographic study.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4D5G
 
 | | Structure of recombinant CDH-H28AN484A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CYCLOHEXANE-1,2-DIONE HYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6J40
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
3HL1
 
 | |
6HTH
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 5 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-[(4-phenylphenyl)carbonylamino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor | | Descriptor: | 1-[5-chloranyl-2-(4-fluoranylphenoxy)phenyl]-~{N}-oxidanyl-1,2,3-triazole-4-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HI2
 
 | | PI3 Kinase Delta in complex with 3{6[(1S,6R)3oxabicyclo[4.1.0]heptan6yl]pyridin2yl}phenol | | Descriptor: | 3-[6-[(1~{S},6~{R})-3-oxabicyclo[4.1.0]heptan-6-yl]pyridin-2-yl]phenol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Summers, D, Peace, S. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A theoretical and experimental investigation into the conformational bias of aryl cyclopropylpyrans, novel bioisosteres for N-aryl morpholines.
to be published
|
|
4CVD
 
 | |
6HSF
 
 | | Crystal structure of Schistosoma mansoni HDAC8 mutant H292M complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
4D5E
 
 | | Crystal Structure of recombinant wildtype CDH | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-03 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3GR3
 
 | |
6HSH
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
4Q9W
 
 | |
4QFR
 
 | | Structure of AMPK in complex with Cl-A769662 activator and STAUROSPORINE inhibitor | | Descriptor: | 2-chloro-4-hydroxy-3-(2'-hydroxybiphenyl-4-yl)-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6KHI
 
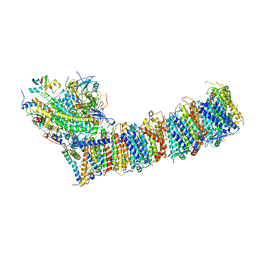 | | Supercomplex for cylic electron transport in cyanobacteria | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
3KL7
 
 | |
4Q9X
 
 | | mTFP* PdCl2 soak | | Descriptor: | CARBONATE ION, CHLORIDE ION, GFP-like fluorescent chromoprotein cFP484, ... | | Authors: | Fischer, J, Quitterer, F, Groll, M, Eppinger, J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | mTFP*: An imperishable and versatile protein host for anchoring diverse ligands and organocatalysts
To be Published
|
|
2GFG
 
 | |
7LAF
 
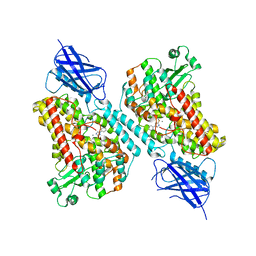 | | 15-lipoxygenase-2 loop mutant bound to imidazole-based inhibitor | | Descriptor: | 3-{[(4-methylphenyl)methyl]sulfanyl}-1-phenyl-1H-1,2,4-triazole, MANGANESE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX15B | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Kinetic and structural investigations of novel inhibitors of human epithelial 15-lipoxygenase-2.
Bioorg.Med.Chem., 46, 2021
|
|
7LY5
 
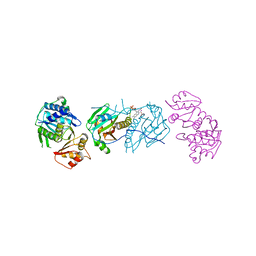 | | Proteolyzed crystal structure of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | BmdB, Bacillamide NRPS, BmdC, ... | | Authors: | Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
5WZM
 
 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZS
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 8 | | Descriptor: | 2-[2-methyl-1-(naphthalen-1-ylmethyl)-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
7LY6
 
 | | Structure of a trans-acting NRPS oxidase, BmdC, involved in bacillamide biosynthesis | | Descriptor: | BmdC, NRPS oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fortinez, C.M, Bloudoff, K, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
5WZV
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Me-indoxam | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[(2-phenylphenyl)methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
