3DY0
 
 | | Crystal Structure of Cleaved PCI Bound to Heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-terminus Plasma serine protease inhibitor, GLYCEROL, ... | | Authors: | Li, W, Huntington, J.A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The heparin binding site of protein C inhibitor is protease-dependent.
J.Biol.Chem., 283, 2008
|
|
2XYL
 
 | | CELLULOMONAS FIMI XYLANASE/CELLULASE COMPLEXED WITH 2-DEOXY-2-FLUORO-XYLOBIOSE | | Descriptor: | BETA-1,4-GLYCANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Monem, V, Birsan, C, Warren, R.A.J, Withers, S.G, Rose, D.R. | | Deposit date: | 1997-11-20 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the cellulose/xylan specificity of the beta-1,4-glycanase cex from Cellulomonas fimi through crystallography and mutation.
Biochemistry, 37, 1998
|
|
3DR5
 
 | | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium target CgR117. | | Descriptor: | Putative O-Methyltransferase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum.
To be Published
|
|
3DXJ
 
 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
3E0M
 
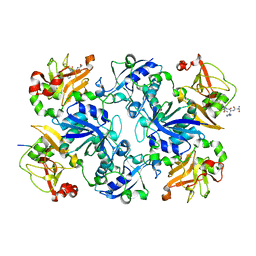 | | Crystal structure of fusion protein of MsrA and MsrB | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB 1, Short peptide SHMAEI | | Authors: | Kim, Y.K, Hwang, K.Y. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Analysis of an MsrA-MsrB Fusion Protein from Streptococcus pneumoniae
Mol.Microbiol., 72, 2009
|
|
3RLA
 
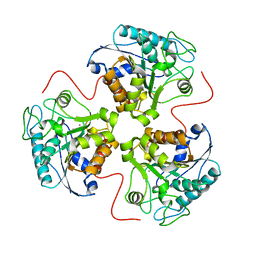 | |
6NLW
 
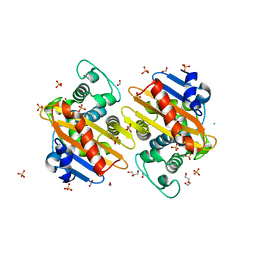 | | The crystal structure of class D carbapenem-hydrolyzing beta-lactamase BlaA from Shewanella oneidensis MR-1 | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-09 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of class D carbapenem-hydrolyzing beta-lactamase BlaA from Shewanella oneidensis MR-1
To Be Published
|
|
2TRH
 
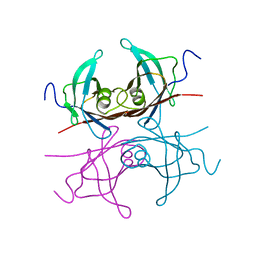 | |
2UCZ
 
 | |
2SRC
 
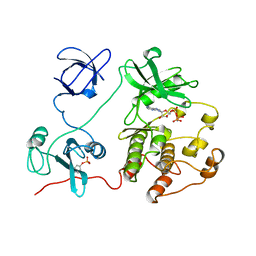 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
2STD
 
 | | SCYTALONE DEHYDRATASE COMPLEXED WITH TIGHT-BINDING INHIBITOR CARPROPAMID | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, SCYTALONE DEHYDRATASE, SULFATE ION | | Authors: | Nakasako, M, Motoyama, T, Kurahashi, Y, Yamaguchi, I. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
Biochemistry, 37, 1998
|
|
2SIV
 
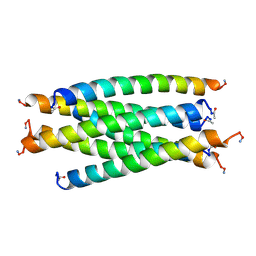 | | SIV GP41 CORE STRUCTURE | | Descriptor: | SIV GP41 GLYCOPROTEIN | | Authors: | Malashkevich, V.N, Chan, D.C, Chutkowski, C.T, Kim, P.S. | | Deposit date: | 1998-06-17 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2TRY
 
 | |
2SOD
 
 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
2TPR
 
 | |
3G61
 
 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
2UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2UPJ
 
 | |
2UUD
 
 | |
3G9H
 
 | | Crystal structure of the C-terminal mu homology domain of Syp1 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
2UVR
 
 | | Crystal structures of mutant Dpo4 DNA polymerases with 8-oxoG containing DNA template-primer constructs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
J.Biol.Chem., 282, 2007
|
|
3G9Q
 
 | | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577A | | Descriptor: | Ferrichrome-binding protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-13 | | Release date: | 2009-03-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577A.
To be Published
|
|
2UUR
 
 | | N-terminal NC4 domain of collagen IX | | Descriptor: | 1,2-ETHANEDIOL, COLLAGEN ALPHA-1(IX) CHAIN, SULFATE ION, ... | | Authors: | Leppanen, V.-M, Tossavainen, H, Permi, P, Lehtio, L, Ronnholm, G, Goldman, A, Kilpelainen, I, Pihlajamaa, T. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Terminal Nc4 Domain of Collagen Ix, a Zinc Binding Member of the Laminin-Neurexin-Sex Hormone Binding Globulin (Lns) Domain Family.
J.Biol.Chem., 282, 2007
|
|
3GAO
 
 | |
3GCB
 
 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
