3PKQ
 
 | | Q83D Variant of S. Enterica RmlA with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3PL1
 
 | | Determination of the crystal structure of the pyrazinamidase from M.tuberculosis : a structure-function analysis for prediction resistance to pyrazinamide. | | Descriptor: | FE (II) ION, PYRAZINAMIDASE/NICOTINAMIDASE PNCA (PZase) | | Authors: | Petrella, S, Gelus-Ziental, N, Mayer, C, Sougakoff, W. | | Deposit date: | 2010-11-12 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Pyrazinamidase of Mycobacterium tuberculosis: Insights into Natural and Acquired Resistance to Pyrazinamide.
Plos One, 6, 2011
|
|
3PSH
 
 | | Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein MolA (MolA bound to Molybdate) | | Descriptor: | MOLYBDATE ION, protein HI_1472 | | Authors: | Tirado-Lee, L, Lee, A, Rees, D.C, Pinkett, H.W. | | Deposit date: | 2010-12-01 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Classification of a Haemophilus influenzae ABC Transporter HI1470/71 through Its Cognate Molybdate Periplasmic Binding Protein, MolA.
Structure, 19, 2011
|
|
3PUM
 
 | | Crystal structure of P domain dimer of Norovirus VA207 | | Descriptor: | Capsid | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3Q4K
 
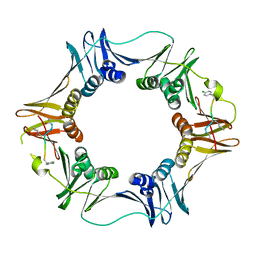 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3PMP
 
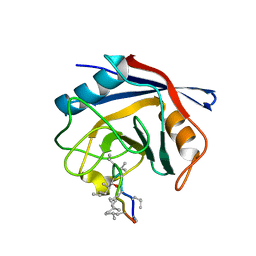 | | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa in complex with Cyclosporin A | | Descriptor: | CYCLOSPORIN A, Cyclophilin A | | Authors: | Monzani, P, Pereira, H.M, Gramacho, K.P, Meirelles, F.V, Oliva, G, Cascardo, J.C.C. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa
To be Published
|
|
3PI2
 
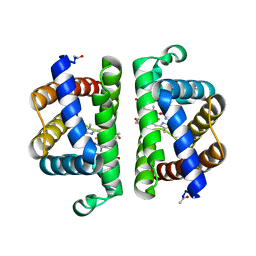 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 8.0 | | Descriptor: | FORMIC ACID, Hemoglobin II, OXYGEN MOLECULE, ... | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
3PJ5
 
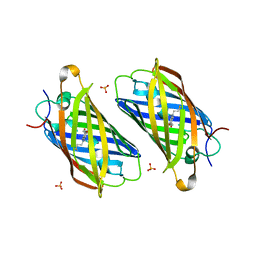 | |
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PQ4
 
 | | Structure of I274C variant of E. coli KatE[] - Images 13-18 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PNL
 
 | | Crystal Structure of E.coli Dha kinase DhaK-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNW
 
 | | Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB | | Descriptor: | FAB heavy chain, FAB light chain, Tudor domain-containing protein 3, ... | | Authors: | Loppnau, P, Tempel, W, Wernimont, A.K, Lam, R, Ravichandran, M, Adams-Cioaba, M.A, Persson, H, Sidhu, S.S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
J.Mol.Biol., 425, 2013
|
|
3PSI
 
 | |
3PT8
 
 | | Structure of HbII-III-CN from Lucina pectinata at pH 5.0 | | Descriptor: | CYANIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gavira, J.A, Ruiz-Martinez, C.R, Nieves-Marrero, C.A, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | New Crystallographic Structure of HbII-III-Oxy and CN forms from Lucina pectinata.
To be Published
|
|
3PUC
 
 | |
3PVS
 
 | | Structure and biochemical activities of Escherichia coli MgsA | | Descriptor: | PHOSPHATE ION, Replication-associated recombination protein A | | Authors: | Page, A.N, George, N.P, Marceau, A.H, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Biochemical Activities of Escherichia coli MgsA.
J.Biol.Chem., 286, 2011
|
|
3PWN
 
 | |
3PV7
 
 | | Crystal structure of NKp30 ligand B7-H6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig-like domain-containing protein DKFZp686O24166/DKFZp686I21167 | | Authors: | Li, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the activating natural killer cell receptor NKp30 bound to its ligand B7-H6 reveals basis for tumor cell recognition in humans
to be published
|
|
3PVD
 
 | | Crystal structure of P domain dimer of Norovirus VA207 complexed with 3'-sialyl-Lewis x tetrasaccharide | | Descriptor: | Capsid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3PX8
 
 | | RTA in complex with 7-carboxy-pterin | | Descriptor: | 2-amino-4-oxo-1,4-dihydropteridine-7-carboxylic acid, Preproricin | | Authors: | Jasheway, K.R, Robertus, J.D. | | Deposit date: | 2010-12-09 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 7-Substituted pterins provide a new direction for ricin A chain inhibitors.
Eur.J.Med.Chem., 46, 2011
|
|
3PXP
 
 | |
3Q0Z
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q1X
 
 | |
3PZH
 
 | |
3Q3L
 
 | | The neutron crystallographic structure of inorganic pyrophosphatase from Thermococcus thioreducens | | Descriptor: | CALCIUM ION, Tt-IPPase | | Authors: | Hughes, R.C, Coates, L, Blakeley, M.P, Tomanicek, S.J, Meehan, E.J, Garcia-Ruiz, J.M, Ng, J.D. | | Deposit date: | 2010-12-22 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (2.5 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
