2G1R
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C Ring | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-{2-[6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2,2-DIMETHYL-3-OXO-2,3-DIHYDRO-4H-1,4-BENZOXAZIN-4-YL]ETHYL}ACETAMIDE, Renin | | Authors: | Holsworth, D.D, Jalaiea, M, Zhanga, E, Mcconnella, P. | | Deposit date: | 2006-02-14 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
1MDZ
 
 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | Descriptor: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1QPY
 
 | | CRYSTAL STRUCTURE OF BACKBONE MODIFIED PNA HEXAMER | | Descriptor: | PEPTIDE NUCLEIC ACID 5'-(*CP1*GPN*TP1*APN*CP1*GPN*LYS)-3' | | Authors: | Haima, G, Rasmussen, H, Schmidt, G, Jensen, D.K, Kastrup, J.S, Stafshede, P.W, Norden, B, Buchardt, O, Nielsen, P.E. | | Deposit date: | 1999-05-14 | | Release date: | 2001-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide Nucleic Acids (PNA) derived from N-(N-methylaminoethyl)glycine. Synthesis, hybridization and structural properties
New J.Chem., 23, 1999
|
|
2R4B
 
 | | ErbB4 kinase domain complexed with a thienopyrimidine inhibitor | | Descriptor: | N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-ethylthieno[3,2-d]pyrimidin-4-amine, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Shewchuk, L.M, Uehling, D.E. | | Deposit date: | 2007-08-31 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3GRS
 
 | |
2SNM
 
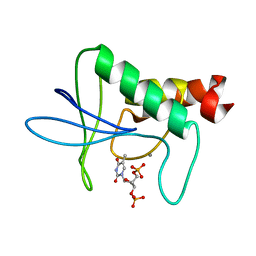 | | IN A STAPHYLOCOCCAL NUCLEASE MUTANT THE SIDE-CHAIN of A LYSINE REPLACING VALINE 66 IS FULLY BURIED IN THE HYDROPHOBIC CORE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Stites, W.E, Gittis, A.G, Lattman, E.E, Shortle, D. | | Deposit date: | 1991-04-03 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core.
J.Mol.Biol., 221, 1991
|
|
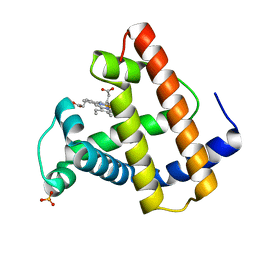
2SPM
 
 | |
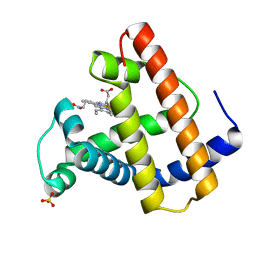
2SPO
 
 | |
1E6D
 
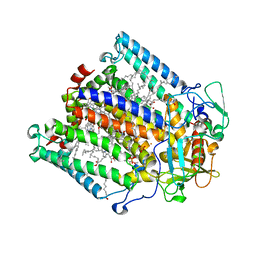 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH TRP M115 REPLACED WITH PHE (CHAIN M, WM115F) PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ridge, J.P, Fyfe, P.K, McAuley, K.E, Van Brederode, M.E, Robert, B, Van Grondelle, R, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2000-08-11 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Examination of How Structural Changes Can Affect the Rate of Electron Transfer in a Mutated Bacterial Photoreaction Centre
Biochem.J., 351, 2000
|
|
284D
 
 | | THE BI-LOOP, A NEW GENERAL FOUR-STRANDED DNA MOTIF | | Descriptor: | BARIUM ION, DNA (5'-CD(*P*AP*TP*TP*CP*AP*TP*TP*C)-3'), SODIUM ION | | Authors: | Salisbury, S.A, Wilson, S.E, Powell, H.R, Kennard, O, Lubini, P, Sheldrick, G.M, Escaja, N, Alazzouzi, E, Grandas, A, Pedroso, E. | | Deposit date: | 1996-09-11 | | Release date: | 1997-06-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The bi-loop, a new general four-stranded DNA motif.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2A0M
 
 | |
2G1O
 
 | |
2G26
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring | | Descriptor: | (5-{[(2R)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)-6-OXOPIPERAZIN-2-YL]METHOXY}-1H-INDOL-1-YL)ACETIC ACID, Renin | | Authors: | Holsworth, D.D, Jalaiea, M, Zhanga, E, Mcconnella, P. | | Deposit date: | 2006-02-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2FS4
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C ring | | Descriptor: | (6R)-6-({[1-(3-HYDROXYPROPYL)-1,7-DIHYDROQUINOLIN-7-YL]OXY}METHYL)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)PIPERAZIN-2-ONE, Renin | | Authors: | Holsworth, D.D, Cai, C, Cheng, X.-M, Cody, W.L, Downing, D.M, Erasga, N, Lee, C, Powell, N.A, Edmunds, J.J, Stier, M, Jalaie, M, Zhang, E, McConnell, P, Ryan, M.J, Bryant, J, Li, T, Kasani, A, Hall, E, Subedi, R, Rahim, M, Maiti, S. | | Deposit date: | 2006-01-20 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2GHC
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2AMV
 
 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
2G24
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring | | Descriptor: | 5-{4-[(3,5-DIFLUOROBENZYL)AMINO]PHENYL}-6-ETHYLPYRIMIDINE-2,4-DIAMINE, Renin | | Authors: | Holsworth, D.D, Jalaiea, M, Zhanga, E, Mcconnella, P. | | Deposit date: | 2006-02-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the C Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
4OGC
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
5A76
 
 | |
1EVJ
 
 | | CRYSTAL STRUCTURE OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE (GFOR) DELTA1-22 S64D | | Descriptor: | GLUCOSE-FRUCTOSE OXIDOREDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lott, J.S, Halbig, D, Baker, H.M, Hardman, M.J, Sprenger, G.A, Baker, E.N. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation.
J.Mol.Biol., 304, 2000
|
|
2IXD
 
 | | Crystal structure of the putative deacetylase BC1534 from Bacillus cereus | | Descriptor: | ACETATE ION, LMBE-RELATED PROTEIN, ZINC ION | | Authors: | Fadouloglou, V.E, Bouriotis, V, Kokkinidis, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bczbp, a Zinc-Binding Protein from Bacillus Cereus
FEBS J., 274, 2007
|
|
1SOQ
 
 | | Crystal structure of the transthyretin mutant A108Y/L110E solved in space group C2 | | Descriptor: | Transthyretin | | Authors: | Hornberg, A, Olofsson, A, Eneqvist, T, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The beta-strand D of transthyretin trapped in two discrete conformations
Biochim.Biophys.Acta, 1700, 2004
|
|
3M3J
 
 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2IZT
 
 | | Structure of casein kinase gamma 3 in complex with inhibitor | | Descriptor: | CASEIN KINASE I ISOFORM GAMMA-3, CHLORIDE ION, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Debreczeni, J.E, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Binding by Casein Kinases
To be Published
|
|
5LCY
 
 | | Formaldehyde-Responsive Regulator FrmR E64H variant from Salmonella enterica serovar Typhimurium | | Descriptor: | Frmr | | Authors: | Pohl, E, Robinson, N, Osman, D, Piergentili, C, Uson, I. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Effectors and Sensory Sites of Formaldehyde-responsive Regulator FrmR and Metal-sensing Variant.
J.Biol.Chem., 291, 2016
|
|
