3KQ0
 
 | | Crystal structure of human alpha1-acid glycoprotein | | Descriptor: | (2R)-2,3-dihydroxypropyl acetate, Alpha-1-acid glycoprotein 1, CHLORIDE ION | | Authors: | Schiefner, A, Schonfeld, D.L, Ravelli, R.B.G, Mueller, U, Skerra, A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin.
J.Mol.Biol., 384, 2008
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
3L27
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L26
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1QKP
 
 | | HIGH RESOLUTION X-RAY STRUCTURE OF AN EARLY INTERMEDIATE IN THE BACTERIORHODOPSIN PHOTOCYCLE | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Edman, K, Nollert, P, Royant, A, Belrhali, H, Pebay-Peyroula, E, Hajdu, J, Neutze, R, Landau, E.M. | | Deposit date: | 1999-07-30 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle.
Nature, 401, 1999
|
|
3L9W
 
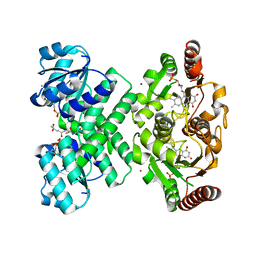 | | KefC C-terminal domain in complex with KefF and GSH | | Descriptor: | ADENOSINE MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Roosild, T.P. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of ligand-gated potassium efflux in bacterial pathogens.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LDY
 
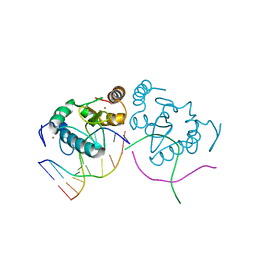 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
8ROL
 
 | |
8ROE
 
 | |
8ROK
 
 | |
8ROH
 
 | | Human cohesin SMC3-HD(EQ)/RAD21-C complex - Apo conformation | | Descriptor: | Double-strand-break repair protein rad21 homolog, GLYCEROL, SULFATE ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROF
 
 | |
8ROC
 
 | |
8ROJ
 
 | | Human cohesin SMC3-HD/RAD21-C complex - ADP-Mg-bound conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
6WN0
 
 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
8ROD
 
 | |
8ROG
 
 | |
8ROI
 
 | | Human cohesin SMC3-HD(EQ)/RAD21-C complex - ADP-bound conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Double-strand-break repair protein rad21 homolog, Structural maintenance of chromosomes protein 3 | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
1AMC
 
 | |
3LK9
 
 | | DNA polymerase beta with a gapped DNA substrate and dTMP(CF2)P(CF2)P | | Descriptor: | 5'-O-[(R)-{[(R)-[difluoro(phosphono)methyl](hydroxy)phosphoryl](difluoro)methyl}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Zibinsky, M, Surya Prakash, G.K, Upton, T.G, Kashemirov, B.A, McKenna, C.E, Oertell, K, Goodman, M.F, Batra, V.K, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2010-01-27 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and biological evaluation of fluorinated deoxynucleotide analogs based on bis-(difluoromethylene)triphosphoric acid.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1EQW
 
 | | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Pesce, A, Battistoni, A, Stroppolo, M.E, Polizio, F, Nardini, M, Kroll, J.S, Langford, P.R, O'Neill, P, Sette, M, Desideri, A, Bolognesi, M. | | Deposit date: | 2000-04-06 | | Release date: | 2000-09-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene.
J.Mol.Biol., 302, 2000
|
|
7LVP
 
 | | Cryptococcus neoformans AIR synthetase | | Descriptor: | 1,2-ETHANEDIOL, AIR synthase, GLYCINE, ... | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
7SWQ
 
 | |
7LVO
 
 | | Cryptococcus neoformans GAR synthetase | | Descriptor: | 1,2-ETHANEDIOL, phosphoribosyl-glycinamide (GAR) synthetase | | Authors: | Chua, S.M.H, Luo, Z, Lim, B.Y.J, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural features of Cryptococcus neoformans bifunctional GAR/AIR synthetase may present novel antifungal drug targets.
J.Biol.Chem., 297, 2021
|
|
3LU8
 
 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
