6HMS
 
 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
6HMF
 
 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) H451 proof-reading deficient variant | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Raia, P, Delarue, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
4J7M
 
 | |
2L2K
 
 | |
4DJJ
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution | | Descriptor: | PIMELIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution
To be Published
|
|
3T5Q
 
 | | 3A structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3'), ... | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TZL
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane
To be Published
|
|
2CFO
 
 | | Non-Discriminating Glutamyl-tRNA Synthetase from Thermosynechococcus elongatus in Complex with Glu | | Descriptor: | GLUTAMIC ACID, GLUTAMYL-TRNA SYNTHETASE | | Authors: | Schulze, J.O, Nickel, D, Schubert, W.-D, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a Non-Discriminating Glutamyl- tRNA Synthetase.
J.Mol.Biol., 361, 2006
|
|
4ERX
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution
To be Published
|
|
7L0Z
 
 | | Spinach variant bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Truong, L, Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2020-12-13 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fluorogenic aptamers resolve the flexibility of RNA junctions using orientation-dependent FRET.
Rna, 27, 2021
|
|
4PBT
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PBR
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-(2-bromoisobutyramido)-phenylalanine (BibaF) | | Descriptor: | 4-[(2-bromo-2-methylpropanoyl)amino]-L-phenylalanine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PBS
 
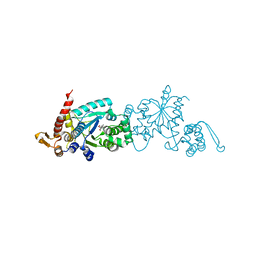 | |
4G0J
 
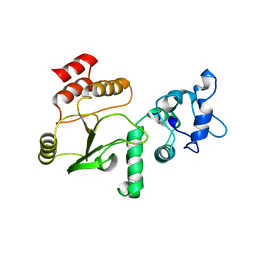 | |
1EHZ
 
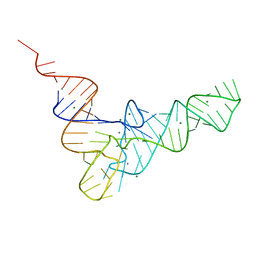 | |
7EIV
 
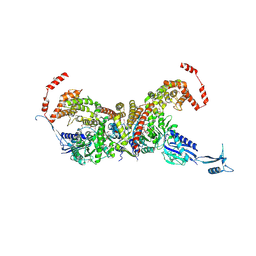 | | heterotetrameric glycyl-tRNA synthetase from Escherichia coli | | Descriptor: | GLYCINE, Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, ... | | Authors: | Ju, Y, Zhou, H. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | X-shaped structure of bacterial heterotetrameric tRNA synthetase suggests cryptic prokaryote functions and a rationale for synthetase classifications.
Nucleic Acids Res., 49, 2021
|
|
3T5N
 
 | | 1.8A crystal structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | NICKEL (II) ION, Nucleoprotein, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3') | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5AF0
 
 | | MAEL domain from Bombyx mori Maelstrom | | Descriptor: | MAELSTROM, ZINC ION | | Authors: | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-01-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
1R1B
 
 | | EPRS SECOND REPEATED ELEMENT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Mirande, M, Guittet, E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1Y42
 
 | | Crystal structure of a C-terminally truncated CYT-18 protein | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase, mitochondrial | | Authors: | Paukstelis, P.J, Coon, R, Madabusi, L, Nowakowski, J, Monzingo, A, Robertus, J, Lambowitz, A.M. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface.
Mol.Cell, 17, 2005
|
|
2EL7
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus
To be published
|
|
8DO6
 
 | | The structure of S. epidermidis Cas10-Csm bound to target RNA | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Paraan, M, Stagg, S.M, Dunkle, J.A. | | Deposit date: | 2022-07-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Plos One, 18, 2023
|
|
1ZFT
 
 | |
1ZFV
 
 | | The structure of an all-RNA minimal Hairpin Ribozyme with Mutation G8A at the cleavage site | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*AP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Wedekind, J.E. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
1ZFX
 
 | | The Structure of a minimal all-RNA Hairpin Ribozyme with the mutant G8U at the cleavage site | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*UP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Wedekind, J.E. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
