6IMC
 
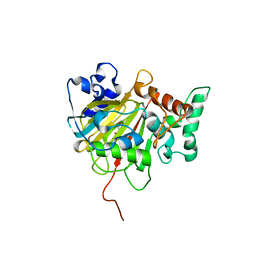 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6OC6
 
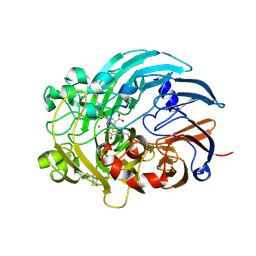 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum and Pyrroloquinoline quinone | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
2MLU
 
 | |
6ALD
 
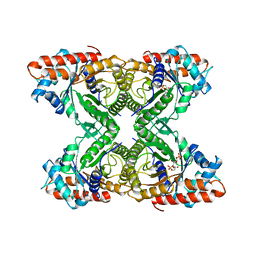 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | Authors: | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
6OYH
 
 | | Crystal structure of MraY bound to carbacaprazamycin | | Descriptor: | (5S)-5'-O-(5-amino-5-deoxy-beta-D-ribofuranosyl)-5'-C-[(2S,5S,6S)-5-carboxy-6-heptadecyl-1,4-dimethyl-3-oxo-1,4-diazepan-2-yl]uridine, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
7K65
 
 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OEL
 
 | | Engineered Fab bound to IL-4 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, Interleukin-4 receptor subunit alpha, ... | | Authors: | Jude, K.M, Moraga, I, Spangler, J.B, Garcia, K.C. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A strategy for the selection of monovalent antibodies that span protein dimer interfaces.
J.Biol.Chem., 294, 2019
|
|
6IRX
 
 | |
6OET
 
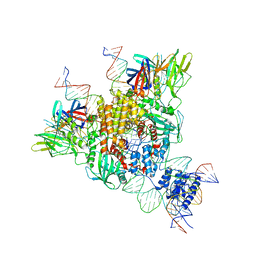 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6AOC
 
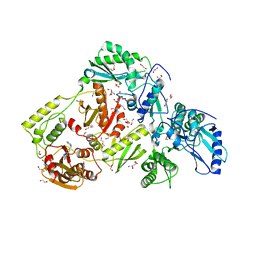 | | Crystal Structure of an N-Hydroxythienopyrimidine-2,4-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-3-hydroxythieno[2,3-d]pyrimidine-2,4(1H,3H)-dione, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-08-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Eur J Med Chem, 141, 2017
|
|
2MLV
 
 | |
6AMO
 
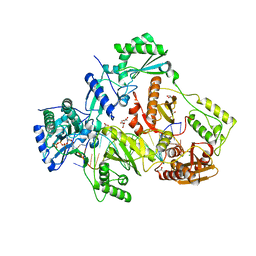 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 7.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-10 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
2YIU
 
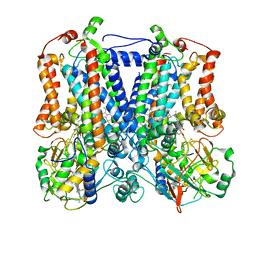 | | X-ray structure of the dimeric cytochrome BC1 complex from the soil bacterium paracoccus denitrificans at 2.7 angstrom resolution | | Descriptor: | CYTOCHROME B, CYTOCHROME C1, HEME PROTEIN, ... | | Authors: | Kleinschroth, T, Castellani, M, Trinh, C.H, Morgner, N, Brutschy, B, Ludwig, B, Hunte, C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure of the Dimeric Cytochrome Bc(1) Complex from the Soil Bacterium Paracoccus Denitrificans at 2.7-A Resolution.
Biochim.Biophys.Acta, 1807, 2011
|
|
6OGI
 
 | | 70S termination complex with RF2 bound to the UAG codon. Rotated ribosome conformation (Structure V) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6IRY
 
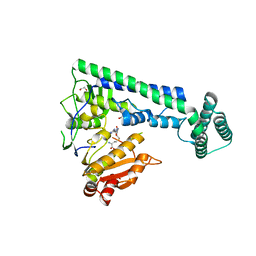 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
2Y6Z
 
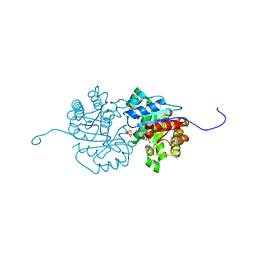 | | Crystallographic structure of GM23 an example of Catalytic migration from TIM to thiamin phosphate synthase. | | Descriptor: | PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, TRIOSE-PHOSPHATE ISOMERASE | | Authors: | Saab-Rincon, G, Olvera, L, Olvera, M, Rudino-Pinera, E, Soberon, X, Morett, E. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Walk between (Beta/Alpha)(8) Barrels: Catalytic Migration from Triosephosphate Isomerase to Thiamin Phosphate Synthase.
J.Mol.Biol., 416, 2012
|
|
6OQC
 
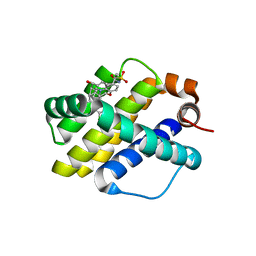 | | Crystal structure of Mcl1 with inhibitor 9 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-10-hydroxy-15-methyl-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
CANCER DISCOV, 8, 2018
|
|
6OQT
 
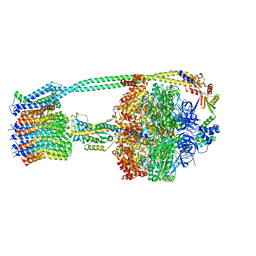 | | E. coli ATP synthase State 1c | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
2MO5
 
 | | hIFABP-oleate complex | | Descriptor: | Fatty acid-binding protein, intestinal, OLEIC ACID | | Authors: | Patil, R, Mohanty, B, Headey, S, Porter, C, Scanlon, M. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of oleate bound human intestinal fatty acid binding protein
To be Published
|
|
7JM4
 
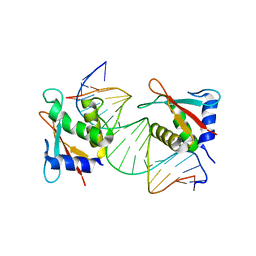 | | IRF Transcription Factor | | Descriptor: | Interferon regulatory factor 4, Interferon-Stimulated Response Elements | | Authors: | Sundararaj, S, Williams, S.J, Casarotto, M.G. | | Deposit date: | 2020-07-31 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural determinants of the IRF4/DNA homodimeric complex.
Nucleic Acids Res., 49, 2021
|
|
6AUO
 
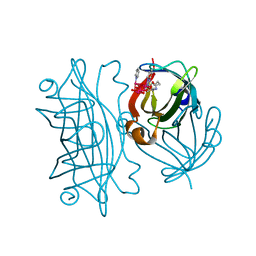 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112F | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)3(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
7K02
 
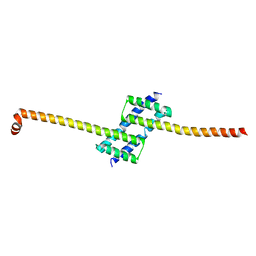 | |
6AUV
 
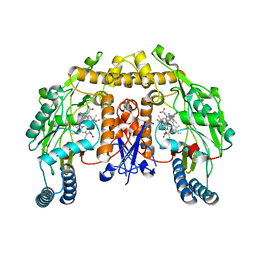 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-Methyl-6-(2-(5-(3-((methylamino)methyl)phenyl)pyridin-3-yl)ethyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-[2-(5-{3-[(methylamino)methyl]phenyl}pyridin-3-yl)ethyl]pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6AUZ
 
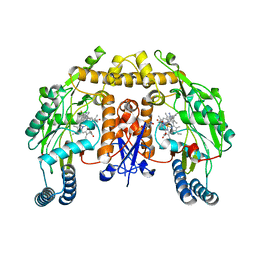 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-Fluoro-5-(3-(methylamino)propyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-fluoro-5-[3-(methylamino)propyl]phenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
