7UWF
 
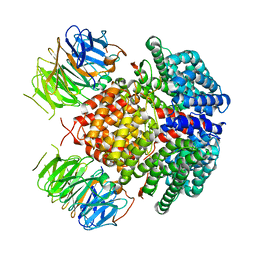 | | Human Rix1 sub-complex scaffold | | Descriptor: | Modulator of non-genomic activity of estrogen receptor, WD repeat-containing protein 18 | | Authors: | Gordon, J, Stanley, R.E. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Nat Commun, 13, 2022
|
|
7VMM
 
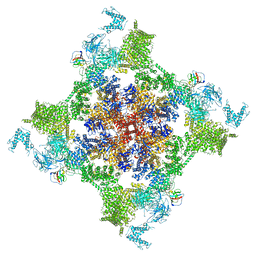 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
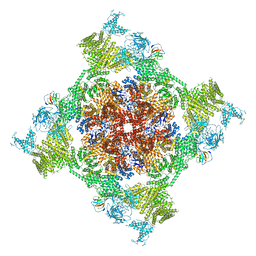 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
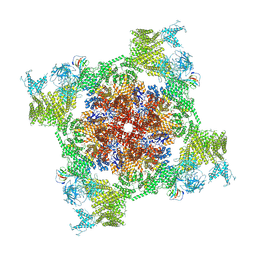 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7XKO
 
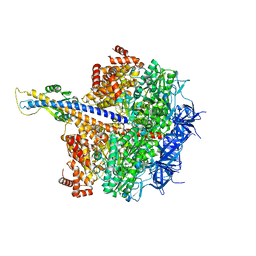 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,nucleotide depeleted | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKQ
 
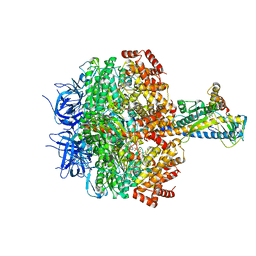 | | F1 domain of FoF1-ATPase with the down form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7Y45
 
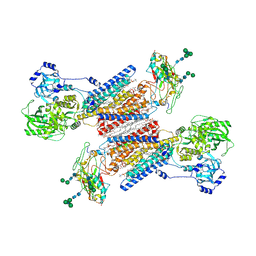 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7XKR
 
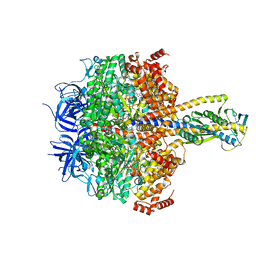 | | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7Y46
 
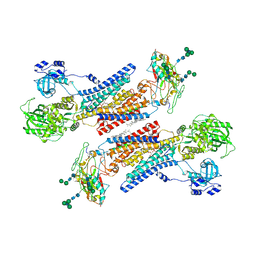 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7XKP
 
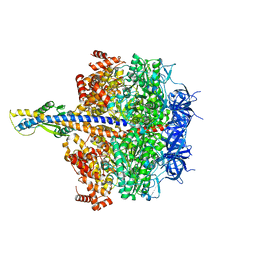 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
6KWV
 
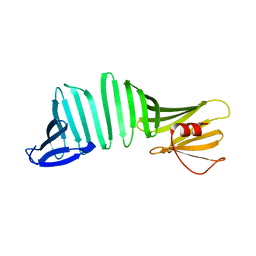 | | A Crystal Structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Kiya, M, Makabe, K. | | Deposit date: | 2019-09-08 | | Release date: | 2020-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
6KX1
 
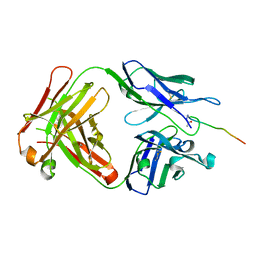 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KXD
 
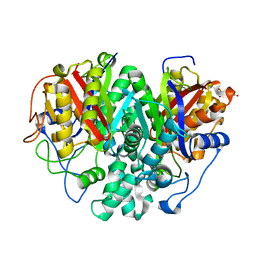 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXN
 
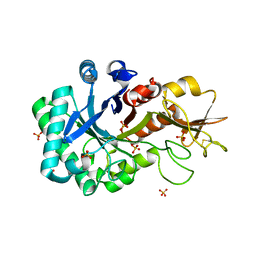 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
6KU9
 
 | |
6KV0
 
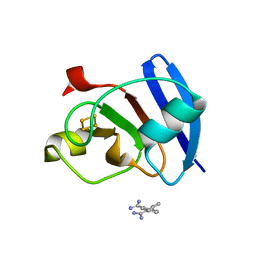 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KV9
 
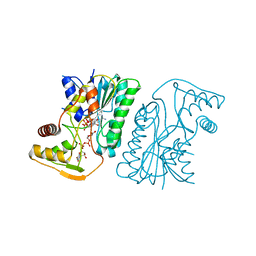 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6KVE
 
 | |
6KXV
 
 | | Crystal structure of a nucleosome containing Leishmania histone H3 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Dacher, M, Taguchi, H, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Incorporation and influence of Leishmania histone H3 in chromatin.
Nucleic Acids Res., 47, 2019
|
|
2HYU
 
 | | Human Annexin A2 with heparin tetrasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
6KWR
 
 | |
2I4Z
 
 | |
