1KG2
 
 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG7
 
 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG5
 
 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1IXR
 
 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
2A5R
 
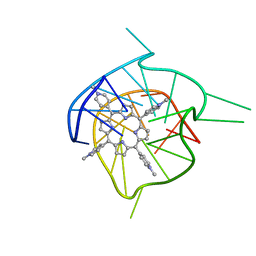 | | Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, C-MYC promoter, NMR, 6 struct. | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
7C7O
 
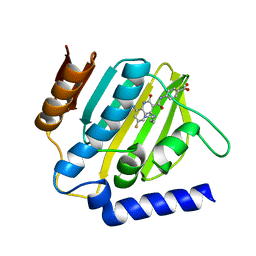 | | Crystal structure of E.coli DNA gyrase B in complex with 6-fluoro-8-(methylamino)-2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[4-(3-azanylpropylamino)-6-fluoranyl-8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Kamitani, M, Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of 8-(methylamino)-2-oxo-1,2-dihydroquinolines as bacterial type II topoisomerase inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
1IXS
 
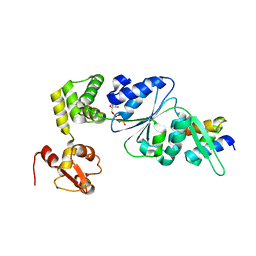 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
5DYM
 
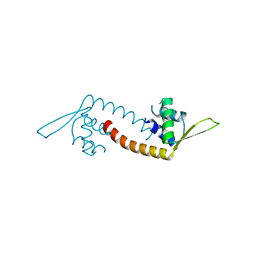 | | Crystal structure of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291 - CdPadR_0991 to 1.89 Angstrom resolution | | Descriptor: | PadR-family transcriptional regulator | | Authors: | Isom, C.E, Karr, E.A, Menon, S.K, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Crystal structure and DNA binding activity of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291.
Bmc Microbiol., 16, 2016
|
|
7C7N
 
 | |
1MX0
 
 | | Structure of topoisomerase subunit | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2002-10-01 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution
Embo J., 22, 2003
|
|
249D
 
 | | STRUCTURAL COMPARISON BETWEEN THE D(CTAG) SEQUENCE IN OLIGONUCLEOTIDES AND TRP AND MET REPRESSOR-OPERATOR COMPLEXES | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*CP*TP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Urpi, L, Tereshko, V, Malinina, L, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1996-02-22 | | Release date: | 1996-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes.
Nat.Struct.Biol., 3, 1996
|
|
6EKR
 
 | |
1IMR
 
 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
7F4N
 
 | | Crystal structure of SAH-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4P
 
 | | Crystal structure of SAM-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4O
 
 | | Crystal structure of MTA1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4Q
 
 | | Crystal structure of SAH-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4L
 
 | | Crystal structure of MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4M
 
 | | Crystal structure of SAM-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
1IMS
 
 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
3THT
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
5AF3
 
 | |
1OLO
 
 | |
