2PLA
 
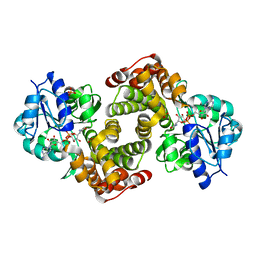 | | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein | | Descriptor: | CHLORIDE ION, Glycerol-3-phosphate dehydrogenase 1-like protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Uppenberg, J, Smee, C, Hozjan, V, Kavanagh, K, Bunkoczi, G, Papagrigoriou, E, Pike, A.C.W, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein.
To be Published
|
|
2PWZ
 
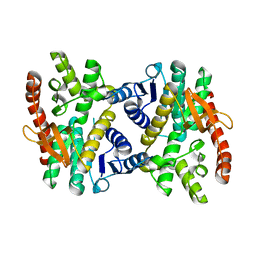 | |
6H98
 
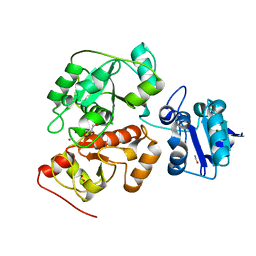 | | Native crystal structure of anaerobic ergothioneine biosynthesis enzyme from Chlorobium limicola. | | Descriptor: | CHLORIDE ION, ETHANOL, FORMIC ACID, ... | | Authors: | Leisinger, F, Burn, R, Meury, M, Lukat, P, Seebeck, F.P. | | Deposit date: | 2018-08-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
2PX4
 
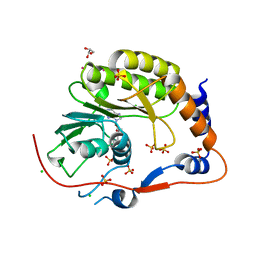 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Monoclinic form 2) | | Descriptor: | CHLORIDE ION, GLYCEROL, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2NVB
 
 | | Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases (ADHs) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Goihberg, E, Tel-Or, S, Peretz, M, Frolow, F, Dym, O, Burstein, Y, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution
Proteins, 72, 2008
|
|
4ACM
 
 | | CDK2 IN COMPLEX WITH 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}-PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2, GLYCEROL | | Authors: | Berg, S, Bhat, R, Anderson, M, Bergh, M, Brassington, C, Hellberg, S, Jerning, E, Hogdin, K, Lo-Alfredsson, Y, Neelissen, J, Nilsson, Y, Ormo, M, Soderman, P, Stanway, J, Tucker, J, von Berg, S, Weigelt, T, Xue, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
2PX5
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Orthorhombic crystal form) | | Descriptor: | Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
8OHS
 
 | |
6H99
 
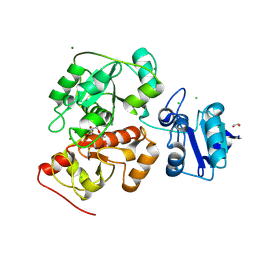 | | Crystal structure of anaerobic ergothioneine biosynthesis enzyme from Chlorobium limicola in persulfide form. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leisinger, F, Burn, R, Meury, M, Lukat, P, Seebeck, F.P. | | Deposit date: | 2018-08-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
2PEG
 
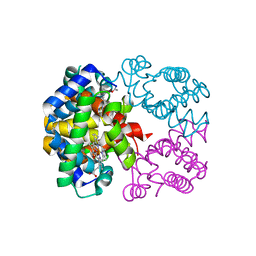 | | Crystal structure of Trematomus bernacchii hemoglobin in a partial hemichrome state | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Franzese, M, Merlino, A, Vitagliano, L, Mazzarella, L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of ferric hemoglobins from three antarctic fish species of the suborder notothenioidei.
Biophys.J., 93, 2007
|
|
2Q7M
 
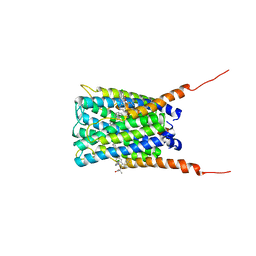 | | Crystal structure of human FLAP with MK-591 | | Descriptor: | 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2PX8
 
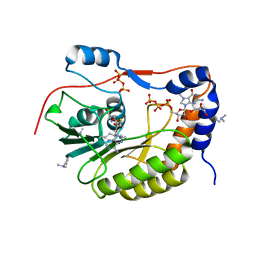 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and 7M-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PXC
 
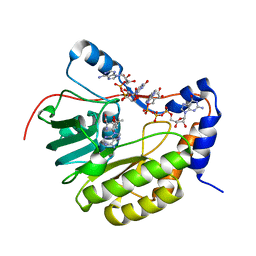 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAM and GTPA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYLMETHIONINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PXA
 
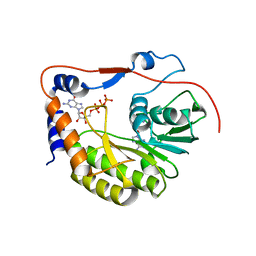 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and GTPG | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PX2
 
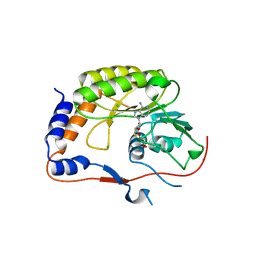 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Monoclinic form 1) | | Descriptor: | CHLORIDE ION, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2NO4
 
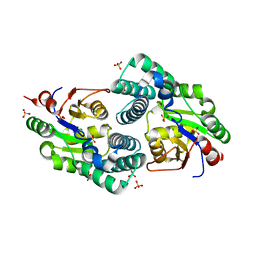 | | Crystal Structure analysis of a Dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2NO5
 
 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | Descriptor: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2R0Z
 
 | | PFA1 FAB complexed with GripI peptide fragment | | Descriptor: | GLYCEROL, GripI peptide fragment, IgG2a Fab fragment heavy chain, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6Z62
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.156 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
4C7A
 
 | | Crystal structure of the Smoothened CRD, selenomethionine-labeled | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
4BS0
 
 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
3EDC
 
 | |
4DFP
 
 | | Crystal structure of the large fragment of DNA Polymerase I from Thermus aqauticus in a ternary complex with 7-(aminopentinyl)-7-deaza-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-5-(5-aminopent-1-yn-1-yl)-7-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 5'-d(AAACGGCGCCGTGGTC)-3', ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
8GDO
 
 | |
