3GIQ
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3FDK
 
 | | Crystal structure of hydrolase DR0930 with promiscuous catalytic activity | | Descriptor: | HYDROLASE DR0930, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, L.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2008-11-25 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3SID
 
 | | Crystal structure of oxidized Symerythrin from Cyanophora paradoxa, azide adduct at 50% occupancy | | Descriptor: | AZIDE ION, FE (III) ION, Symerythrin | | Authors: | Cooley, R.B, Arp, D.J, Karplus, P.A. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Symerythrin structures at atomic resolution and the origins of rubrerythrins and the ferritin-like superfamily.
J.Mol.Biol., 413, 2011
|
|
1TZ6
 
 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica complexed with aminoimidazole riboside and ATP analog | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
2KQN
 
 | |
6UID
 
 | |
1MHH
 
 | |
5ENP
 
 | | MBX2931 bound structure of bacterial efflux pump. | | Descriptor: | 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)piperazin-1-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2FQ6
 
 | |
5ENQ
 
 | | MBX3132 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{R})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]ethanamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENT
 
 | | Minocycline bound structure of bacterial efflux pump. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENR
 
 | | MBX3135 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]prop-2-enamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8R1X
 
 | |
5ENS
 
 | | Rhodamine bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, RHODAMINE 6G | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQ8
 
 | |
5ENO
 
 | | MBX2319 bound structure of bacterial efflux pump. | | Descriptor: | 3,3-dimethyl-8-morpholin-4-yl-6-(2-phenylethylsulfanyl)-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQA
 
 | |
6UIE
 
 | | Structure of the cytoplasmic domain of the T3SS sorting platform protein PscK from P. aeruginosa | | Descriptor: | CHLORIDE ION, Type III export protein PscK | | Authors: | Muthuramalingam, M, Lovell, S, Battaile, K.P, Picking, W.D. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structures of SctK and SctD from Pseudomonas aeruginosa Reveal the Interface of the Type III Secretion System Basal Body and Sorting Platform.
J.Mol.Biol., 432, 2020
|
|
5EN5
 
 | | Apo structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQ7
 
 | |
1SCQ
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
5UKW
 
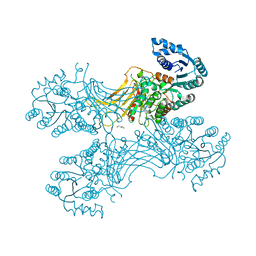 | |
4JM3
 
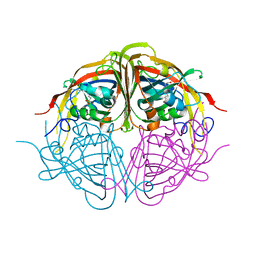 | | Enduracididine Biosynthesis Enzyme MppR with HEPES Buffer Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative uncharacterized protein mppR | | Authors: | Silvaggi, N.R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Characterization of MppR, an Enduracididine Biosynthetic Enzyme from Streptomyces hygroscopicus: Functional Diversity in the Acetoacetate Decarboxylase-like Superfamily.
Biochemistry, 52, 2013
|
|
4JME
 
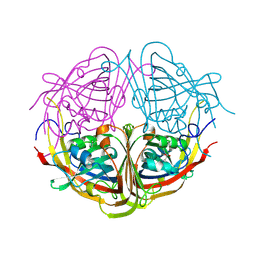 | | Enduracididine biosynthesis enzyme MppR complexed with 2-keto-enduracididine | | Descriptor: | (4R)-4-(2-carboxyethyl)imidazolidin-2-iminium, Putative uncharacterized protein mppR | | Authors: | Silvaggi, N.R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of MppR, an Enduracididine Biosynthetic Enzyme from Streptomyces hygroscopicus: Functional Diversity in the Acetoacetate Decarboxylase-like Superfamily.
Biochemistry, 52, 2013
|
|
2HIN
 
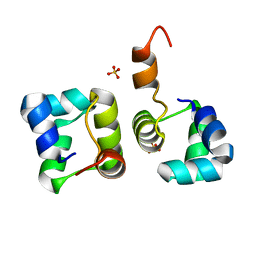 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
