3VAJ
 
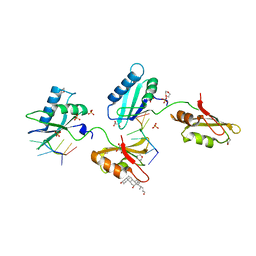 | | Structure of U2AF65 variant with BrU5C6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*CP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
2JSQ
 
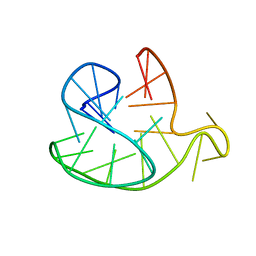 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 15BrG, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
3VAI
 
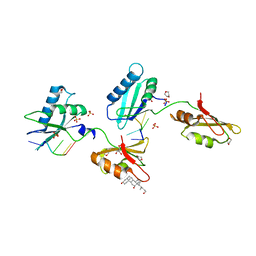 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
8U10
 
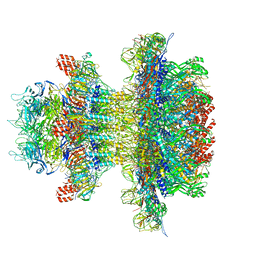 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8U11
 
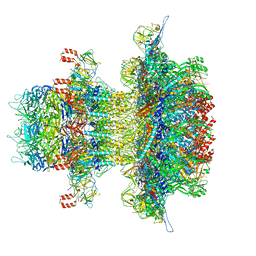 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
7LL0
 
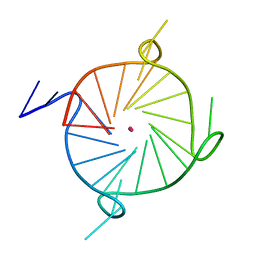 | |
1NO4
 
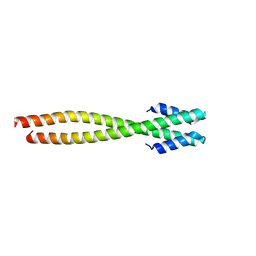 | | Crystal Structure of the pre-assembly scaffolding protein gp7 from the double-stranded DNA bacteriophage phi29 | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.A.L, McMurray, C.T, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2003-01-15 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
2JSK
 
 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, 16 G Form 1, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-07 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
2JSM
 
 | | MONOMERIC HUMAN TELOMERE DNA TETRAPLEX WITH 3+1 STRAND FOLD TOPOLOGY, TWO EDGEWISE LOOPS AND DOUBLE-CHAIN REVERSAL LOOP, NMR, 10 STRUCTURES, Form 1 Natural | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution
Nucleic Acids Res., 35, 2007
|
|
3UYB
 
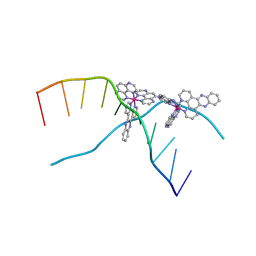 | | X-ray crystal structure of the ruthenium complex [Ru(tap)2(dppz)]2+ bound to d(TCGGTACCGA) | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', BARIUM ION, Ru(tap)2(dppz) complex | | Authors: | Niyazi, H.N, Texeira, S.C.M, Mitchell, E.P, Cardin, J.C, Forsyth, V.T. | | Deposit date: | 2011-12-06 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lambda-[Ru(TAP)2(dppz)]2+ with d(TCGGTACCGA)2 displaying kinking of DNA via semiintercalation.
To be Published
|
|
7MC3
 
 | | Solution structure of Miz-1 zinc finger 12 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MC1
 
 | | Solution structure of Miz-1 Zinc finger 10 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MC2
 
 | | Solution structure of Miz-1 Zinc finger 11 H586Y | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
481D
 
 | |
1H3G
 
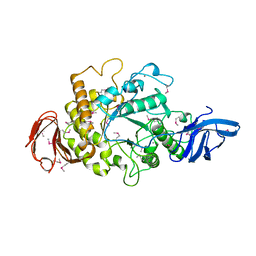 | | Cyclomaltodextrinase from Flavobacterium sp. No. 92: from DNA sequence to protein structure | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase | | Authors: | Fritzsche, H.B, Schwede, T, Jelakovic, S, Schulz, G.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-08-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Covalent and Three-Dimensional Structure of the Cyclodextrinase from Flavobacterium Sp. No. 92.
Eur.J.Biochem., 270, 2003
|
|
8HIS
 
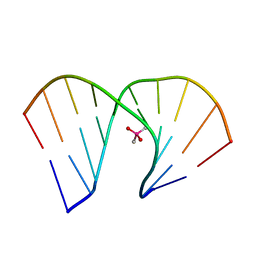 | | Crystal structure of DNA decamer containing GuNA[Me,tBu] | | Descriptor: | CACODYLIC ACID, DNA (5'-D(*GP*CP*GP*TP*AP*(LR6)P*AP*CP*GP*C)-3') | | Authors: | Aoyama, H, Obika, S, Yamaguchi, T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanism of the extremely high duplex-forming ability of oligonucleotides modified with N-tert-butylguanidine- or N-tert-butyl-N'- methylguanidine-bridged nucleic acids.
Nucleic Acids Res., 51, 2023
|
|
8IJC
 
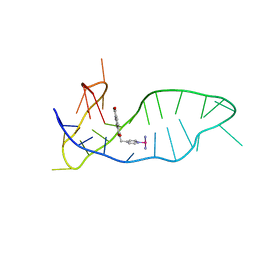 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2I1Q
 
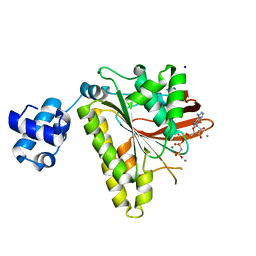 | | RadA Recombinase in complex with Calcium | | Descriptor: | CALCIUM ION, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Qian, X, He, Y, Ma, X, Fodje, M.N, Grochulski, P, Luo, Y. | | Deposit date: | 2006-08-14 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium Stiffens Archaeal Rad51 Recombinase from Methanococcus voltae for Homologous Recombination.
J.Biol.Chem., 281, 2006
|
|
3SC8
 
 | | Crystal structure of an intramolecular human telomeric DNA G-quadruplex bound by the naphthalene diimide BMSG-SH-3 | | Descriptor: | 2,7-bis[3-(4-methylpiperazin-1-yl)propyl]-4,9-bis{[3-(4-methylpiperazin-1-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, Human telomeric repeat sequence, POTASSIUM ION | | Authors: | Collie, G.W, Promontorio, R, Parkinson, G.N. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural basis for telomeric g-quadruplex targeting by naphthalene diimide ligands.
J.Am.Chem.Soc., 134, 2012
|
|
1QZR
 
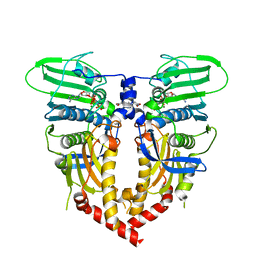 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF SACCHAROMYCES CEREVISIAE TOPOISOMERASE II BOUND TO ICRF-187 (DEXRAZOXANE) | | Descriptor: | (S)-4,4'-(1-METHYL-1,2-ETHANEDIYL)BIS-2,6-PIPERAZINEDIONE, DNA topoisomerase II, MAGNESIUM ION, ... | | Authors: | Classen, S, Olland, S, Berger, J.M. | | Deposit date: | 2003-09-17 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4K4O
 
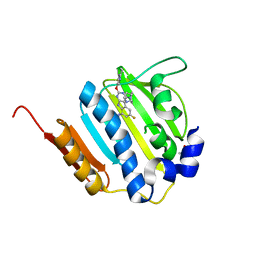 | | The DNA Gyrase B ATP binding domain of Enterococcus faecalis in complex with a small molecule inhibitor | | Descriptor: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | Deposit date: | 2013-04-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A new class of type IIA topoisomerase inhibitors with broad-spectrum antibacterial
activity
To be Published
|
|
7F4N
 
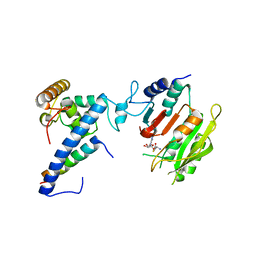 | | Crystal structure of SAH-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4P
 
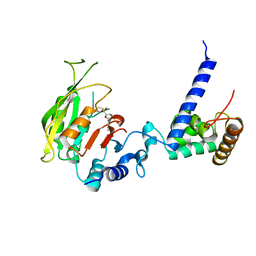 | | Crystal structure of SAM-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4O
 
 | | Crystal structure of MTA1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4Q
 
 | | Crystal structure of SAH-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
