8BH5
 
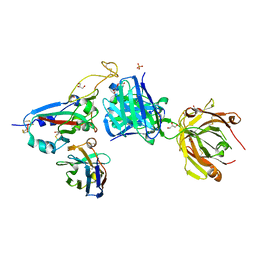 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
1UCI
 
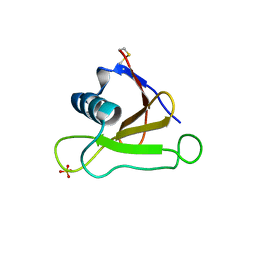 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
8B7Z
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Palm, G.J, Hinrichs, W. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
1U0G
 
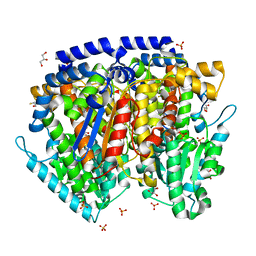 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
8B02
 
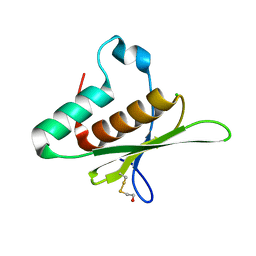 | | Crystal structure of the dsRBD domain of tRNA-dihydrouridine(20) synthase from Amphimedon queenslandica | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DRBM domain-containing protein, ... | | Authors: | Pecqueur, L, Faivre, B, Hamdane, D. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family.
Biomolecules, 12, 2022
|
|
1UCR
 
 | | Three-dimensional crystal structure of dissimilatory sulfite reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Mizuno, N, Voordouw, G, Miki, K, Sarai, A, Higuchi, Y. | | Deposit date: | 2003-04-18 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Dissimilatory Sulfite Reductase D (DsrD) Protein-Possible Interaction with B- and Z-DNA by Its Winged-Helix Motif
STRUCTURE, 11, 2003
|
|
3S9T
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-{[(4,6-dimethyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-chloro-5-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
8BD1
 
 | |
1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UDV
 
 | | Crystal structure of the hyperthermophilic archaeal dna-binding protein Sso10b2 at 1.85 A | | Descriptor: | DNA binding protein SSO10b, ZINC ION | | Authors: | Chou, C.-C, Lin, T.-W, Chen, C.-Y, Wang, A.H.J. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms
J.BACTERIOL., 185, 2003
|
|
8BAO
 
 | |
8BHZ
 
 | |
1UE3
 
 | | Crystal structure of d(GCGAAAGC) containing hexaamminecobalt | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6EZT
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi | | Descriptor: | Beta-N-acetylglucosaminidase Nag2, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
3SA2
 
 | |
3SAA
 
 | | Crystal structure of Wild-type HIV-1 protease in complex With AF77 | | Descriptor: | N-[(2S,3R)-4-{(cyclohexylmethyl)[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-3-hydroxybenzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
1U14
 
 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
8B2J
 
 | | Crystal structure of human STING in complex with ADU-S100 | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, CALCIUM ION, SODIUM ION, ... | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L. | | Deposit date: | 2022-09-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Crystal structure of human STING in complex with ADU-S100
To Be Published
|
|
8BBP
 
 | |
1U17
 
 | |
1U1I
 
 | | Myo-inositol phosphate synthase mIPS from A. fulgidus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stieglitz, K.A, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2004-07-15 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaching for Mechanistic Consensus Across Life Kingdoms: Structure and Insights into Catalysis of the myo-Inositol-1-phosphate Synthase (mIPS) from Archaeoglobus fulgidus
Biochemistry, 44, 2005
|
|
1UEA
 
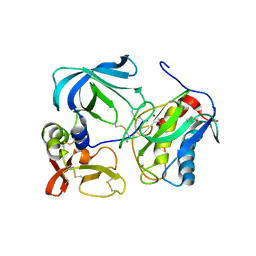 | | MMP-3/TIMP-1 COMPLEX | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-3, TISSUE INHIBITOR OF METALLOPROTEINASE-1, ... | | Authors: | Bode, W, Maskos, K, Gomis-Rueth, F.-X, Nagase, H. | | Deposit date: | 1997-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1.
Nature, 389, 1997
|
|
8BF3
 
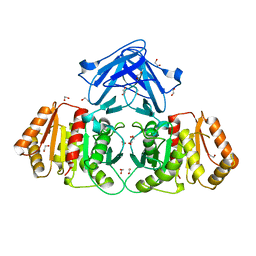 | |
8BLU
 
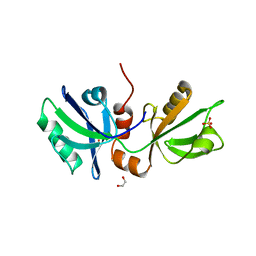 | | The PDZ domains of human SDCBP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP
To Be Published
|
|
3SB6
 
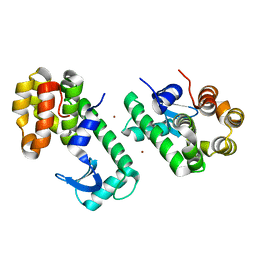 | | Cu-mediated Dimer of T4 Lysozyme D61H/K65H/R76H/R80H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
