3ST8
 
 | |
3SPU
 
 | | apo NDM-1 Crystal Structure | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2011-07-03 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance
Protein Sci., 20, 2011
|
|
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SQD
 
 | |
3SV5
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3SQF
 
 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SQN
 
 | | Putative Mga family transcriptional regulator from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Putative Mga family transcriptional regulator from Enterococcus faecalis.
To be Published
|
|
3SWF
 
 | | CNGA1 621-690 containing CLZ domain | | Descriptor: | ZINC ION, cGMP-gated cation channel alpha-1 | | Authors: | Shuart, N.G, Haitin, Y, Camp, S.S, Black, K.D, Zagotta, W.N. | | Deposit date: | 2011-07-13 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels.
Nat Commun, 2, 2011
|
|
3SR9
 
 | | Crystal structure of mouse PTPsigma | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Wang, J, Hou, L, Li, J, Ding, J. | | Deposit date: | 2011-07-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the homology and differences between mouse protein tyrosine phosphatase-sigma and human protein tyrosine phosphatase-sigma
Acta Biochim.Biophys.Sin., 43, 2011
|
|
3SWK
 
 | |
3SX0
 
 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
3SUF
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SXA
 
 | | Crystal structure of ABBB+UDP+Gal with Glycerol as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Histo-blood group ABO system transferase, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SXG
 
 | | Crystal structure of AAAA+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SV7
 
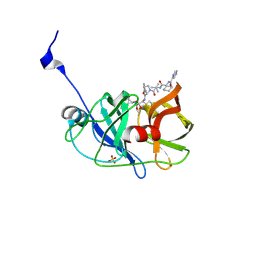 | | Crystal structure of NS3/4A protease variant R155K in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SZY
 
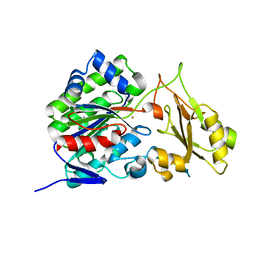 | |
3SYA
 
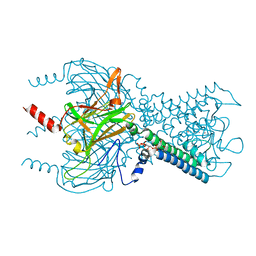 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with sodium and PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-16 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
3T3Y
 
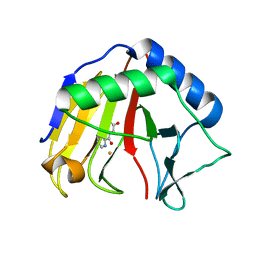 | |
3T43
 
 | |
3T46
 
 | |
3T49
 
 | |
3T07
 
 | | Crystal structure of S. aureus Pyruvate Kinase in complex with a naturally occurring bis-indole alkaloid | | Descriptor: | (3S,5R)-3,5-bis(6-bromo-1H-indol-3-yl)piperazin-2-one, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities.
J.Biol.Chem., 286, 2011
|
|
3T5P
 
 | | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | BmrU protein, MAGNESIUM ION | | Authors: | Hou, J, Zheng, H, Chruszcz, M, Cooper, D.R, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
To be Published
|
|
3T2A
 
 | | TMAO-grown cubic insulin (porcine) | | Descriptor: | Insulin A chain, Insulin B chain, trimethylamine oxide | | Authors: | Cahn, J, Venkat, M, Marshall, H, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3T8O
 
 | | Rhodopsin kinase (GRK1) L166K mutant at 2.5A resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tesmer, J.J.G, Singh, P, Nance, M.R. | | Deposit date: | 2011-08-01 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a monomeric variant of rhodopsin kinase at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
